Two-Step CRISPR Enables Full-Length Mouse Gene Humanisation
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Full-length gene-humanised mice – created by completely replacing mouse genomic loci with human equivalents – offer substantial advantages for studying human gene function in vivo. Unlike transgenic approaches that merely add human genes whilst retaining mouse counterparts, or partial humanisations targeting individual exons, complete locus replacement preserves the entire regulatory landscape, including promoters, enhancers, and untranslated regions that govern tissue-specific expression patterns. However, reliably humanising large genomic regions has remained technically challenging due to limited DNA insert capacities in conventional targeting vectors, complex protocols requiring specialised materials, and poor homologous recombination efficiency in embryonic stem cells.
“This study introduces a streamlined approach that enables full-length gene humanisation through two sequential CRISPR-assisted homologous recombination steps in embryonic stem cells”Jumpei Taguchi et al.
The TECHNO method addresses these limitations through two sequential CRISPR-Cas9-assisted homologous recombination steps in mouse embryonic stem cells (see Figure 1). In the first step, researchers remove the target mouse locus using CRISPR-Cas9 ribonucleoproteins and simultaneously integrate homology arms flanking the corresponding human gene sequence, along with a neomycin resistance cassette for selection.
»This method supports targeted knock-in of genomic fragments (>200 kbp) and is applicable across multiple mouse strains,« the authors note in their Nature Communications paper.
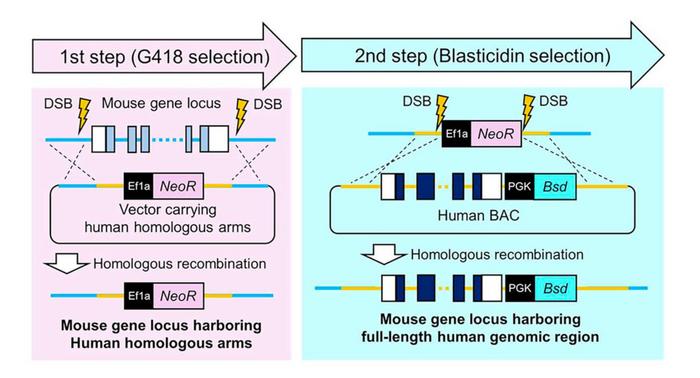
The second step introduces the full-length human genomic fragment – delivered via bacterial artificial chromosome vectors that can accommodate DNA inserts exceeding 200 kilobases – through homologous recombination with the previously inserted targeting sequences. Critically, the approach relies exclusively on standard molecular biology reagents and commercially available BAC resources, theoretically enabling humanisation of 93 per cent of human genes that fall within the approximately 200-kilobase capacity of BAC vectors.
The research team validated their method by humanising three distinct genomic loci of varying sizes and functional importance. Humanisation of the c-Kit locus (approximately 100 kilobases) achieved 11.1 per cent knock-in efficiency using three-kilobase homology arms – substantially higher than the 0.1 to 0.5 per cent efficiency reported for previous large-fragment targeting approaches. RNA sequencing analysis of spermatogonia from homozygous c-Kit-humanised mice revealed expression of all 21 human KIT exons with splicing patterns matching those observed in human cells.
Immunostaining confirmed human c-KIT protein expression in cerebellar molecular layers, pulmonary macrophages, and kidney collecting ducts, faithfully recapitulating the organ-specific distribution documented in human tissues. Whilst some homozygous mice exhibited white coat spotting and reduced testis weights, they survived to adulthood and produced viable offspring through in vitro fertilisation, demonstrating that the humanised allele could largely compensate for essential c-Kit functions in haematopoiesis, spermatogenesis, and survival (see Figure 2).
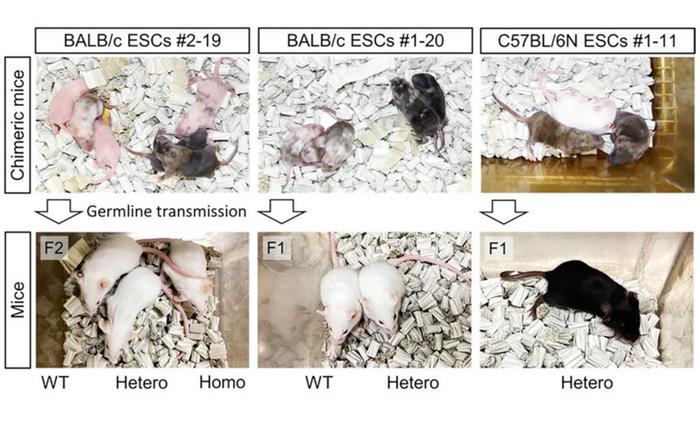
To test the method's scalability, the team successfully replaced the single-gene mouse Apobec3 locus with the human APOBEC3 gene cluster spanning more than 205 kilobases and containing seven tandemly arranged genes (APOBEC3A, B, C, D, F, G, and H). Knock-in efficiencies reached 15.2 per cent in BALB/c embryonic stem cells and 10.6 per cent in C57BL/6N cells, confirming applicability across multiple mouse strains. RNA sequencing of lungs and spleens from Apobec3-humanised mice showed expression of all seven human APOBEC3 genes, with expression profiles correlating significantly with those in human tissues.
Quantitative PCR showed high expression in the spleen and lung, but low expression in skeletal muscle and cerebrum, mirroring the organ-specific patterns observed in humans. Western blot analysis detected APOBEC3B, C, F, G, and H proteins in humanised mouse leukocytes, though APOBEC3A protein was not detected despite mRNA expression.
“H3K9me3, which is deposited shortly after fertilization, is required for the subsequent de novo DNA methylation at the H19-DMR”Takuro Horii et al.
The platform's utility for disease modelling was demonstrated by humanising the X-linked Cybb gene (approximately 55 kilobases), which encodes the NADPH oxidase component responsible for reactive oxygen species production in phagocytes. Following successful humanisation of the Cybb locus, the researchers introduced two chronic granulomatous disease-associated mutations (T458G and A461Δ) into the human CYBB allele via additional genome editing.
Upon phorbol myristate acetate stimulation, granulocytes from wild-type and humanised mice carrying the normal human allele generated comparable reactive oxygen species levels, whereas cells from mice carrying the mutant allele failed to increase reactive oxygen species production – precisely recapitulating the biochemical defect underlying chronic granulomatous disease in human patients.
»Overall, these results demonstrate that our method enables not only FL-GH of individual loci but also precise modelling of human genetic diseases in vivo by introducing disease-associated mutations into humanised alleles,« the authors conclude.
The authors acknowledge that whilst their approach captures complete gene structures including proximal regulatory regions, it may not encompass all distal regulatory elements, such as distant enhancers. They note that some aspects of gene expression differed between humanised mice and humans, exemplified by reduced APOBEC3A expression and the absence of detectable APOBEC3A protein in humanised mouse leukocytes. These discrepancies likely reflect fundamental interspecies differences in transcriptional regulation that cannot be overcome through simple locus replacement. Additionally, large-scale genomic humanisation can potentially influence neighbouring gene expression, as evidenced by reduced Cbx6 mRNA levels in lungs of APOBEC3-humanised mice.
The study was conducted by Jumpei Taguchi and Manabu Ozawa at the University of Tokyo. It was published in Nature Communications on 14 January 2026.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







