Breaking the PAM Barrier – Cas9 Variants With Genome-Wide Targeting Ability

The latest work from Dr. Kleinstiver’s lab has uncovered two novel and exciting Cas9 variants that together allow precise targeting nearly anywhere in the human genome. The ability to edit the entire genome is a major advance in the CRISPR field, and raises hopes of one day curing many genetic diseases with CRISPR-Cas gene therapy.
»These new proteins mean that we can now walk almost every single base pair in the genome and make a DNA double strand break wherever we want. That was not possible before«, says Assistant Professor Ben Kleinstiver, Ph.D., who heads up the lab at Massachusetts General Hospital and Harvard Medical School.
In their study that was recently published in Science, the researchers also showed that the new Cas9 variants could generate mutations known to protect individuals against human diseases. These genetic variants were located in previously inaccessible regions of the genome, which is an important advance for CRISPR-based therapeutics.
I recently caught up with Ben Kleinstiver and the article’s first-author Russell Walton to hear more about the exciting new results.
For several years, the Kleinstiver lab has focused on building and improving CRISPR-Cas gene editing systems for research and clinical use. »The possibility to edit anywhere in the genome would mean that diseases caused by previously inaccessible single point mutations e.g., some types of blindness, may be cured by CRISPR«, explained Ben Kleinstiver when outlining the motivation for the latest work.
The lab actually studies many different Cas enzymes, but the latest work focused on the prototypical and best-studied Cas9 from Streptococcus pyogenes (SpCas9), which requires a 5’ NGG PAM site for activity. A PAM (protospacer adjacent motif) is a short region of DNA that is located immediately after a target site. Each Cas endonuclease requires the presence of a specific PAM to make a cut in the target DNA sequence (see CRISPR-Cas9 Overview Box).
CRISPR-Cas9 Overview
Cas endonucleases evolved as part of an ancient bacterial immune defence against bacteriophages. These enzymes identify and cut invading viral or plasmid DNA from bacterial genomes but only when a specific short DNA sequence is present in the genome nearby. This sequence is known as a PAM (protospacer adjacent motif) and it helps bacteria to recognise self from non-self. Each Cas enzyme requires a specific PAM; some are more stringent than others. From an evolutionary perspective, PAMs protect bacterial genome integrity, but in applications that exploit the gene editing capabilities of the CRISPR-Cas systems, the PAM requirement presents a major bottleneck i.e. only genomic regions with a suitable PAM nearby are accessible for editing.
Previous work in this lab and elsewhere revealed Cas9 variants that tolerate either single nucleotide changes in the 3rd PAM position or recognise an increased number of PAMs, most notably SpCas9-NG, which until recently exhibited the greatest targeting range of any known Cas9 endonuclease. However, even with the increased targeting ranges offered by previous variants, about 75 % of the human genome remained inaccessible to Cas9 endonucleases. From a clinical perspective, this meant that many genetic diseases were not yet amenable to gene editing.
Mutating Cas9 to Relax PAM Stringency
To greatly expand the targeting capacity of SpCas9, the group set out to relax PAM stringency more than had been done before. They started out with one of their earlier mutants SpCas9-VRQR, which can target many non-canonical PAMs including those with variable bases in the 3rd PAM position. The mutations in VRQR occur in its PAM-interacting (PI) domain, which is the domain that helps Cas enzymes to recognise their preferred PAM sequence near potential target sites.
Using VRQR as a scaffold, the group engineered hundreds of new PI domain mutants that they assessed for PAM specificity using a new high-throughput PAM determination assay (HT-PAMDA). This method is an improvement over previous lower-throughput iterations of this approach.
Most studies to date have focused on handfuls of Cas variants that were assayed as purified recombinantly-expressed proteins, or required expression in a heterologous host. But here, hundreds of new mutants were engineered and characterised in parallel. This was a lot of work, and it was only possible thanks to the HT-PAMDA, developed in the Kleinstiver lab (see High-Throughput PAM Determination Assay Box).
A New Method in the Cas Engineering Toolbox
Speaking about the efforts involved in developing the HT-PAMDA, Russell Walton, who carried out most of the experiments, says »A real challenge was to get to a point where the results we obtained in the assay reliably represented what we observed with the same Cas9 variants in human cells.«
The HT-PAMDA is based on the expression of CRISPR nucleases in cell lysates rather than using purified proteins, and has the capacity to analyse more than 100 variants in parallel. Importantly, it also increases translatability to genome editing experiments because the results mimic what goes on in a cellular genome editing context.
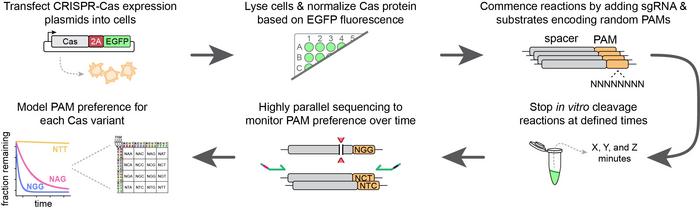
High-Throughput PAM Determination Assay
Cas9 variants are expressed in the human HEK293T cell line in 24- or 96-well plates. Once Cas expression occurs, the cells are gently lysed to release the proteins. A fluorescent label (EGFP) that is translationally de-coupled from each Cas variant, but expressed in approximately equimolar quantities, allows the researcher to normalise the Cas protein concentration so that each variant is assayed at an equal concentration. sgRNAs and a pool of DNA substrates containing random PAM sequences are then added and the reactions are stopped at various time intervals. Whatever is left of the DNA substrates after cleavage is then sequenced and this information is used to infer the PAM specificity and activity of each Cas variant.
Breaking PAM Barriers One Mutant at a Time
Using the PAMDA assay and cellular CRISPR assays with a panel of defined target sites, the group identified SpG as a new variant whose PAM preference was relaxed to a single NGN nucleotide (meaning a glutamine flanked by any other two bases). SpG exhibited even and robust targeting of NGA, NGC, NGG, and NGT PAMs, as well as precise base editing activities across all NGN PAMs.
SpG got the lab excited but it didn’t stop there. »Looking at crystal structure models of how we thought the SpG variant now interacts with DNA, we saw no structural reason for why it shouldn't be able to recognise other sequences«, says Russell, who then explained how they decided to use SpG as a scaffold for further mutations. Guided by the crystal structure of SpCas9, many additional substitutions were tested until the group eventually ended up with a near-PAMless variant that they call SpRY.
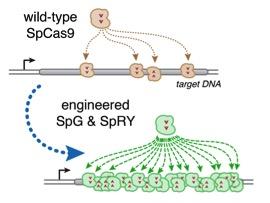
Together, SpG and SpRY enable unconstrained targeting using CRISPR-Cas9 nucleases across nearly the entire genome and with single base pair precision (Figure 1).
Reflecting on the breakthrough, Ben highlights the importance of the PAMDA, saying: »We could only do this work because we could understand the properties of each mutant along the way. The systems, like HT-PAMDA, enabled us to appropriately scale the engineering. You can have as many mutants as you want, but if you don't have the methods to characterise and understand them, you will never really know what they are capable of.«
Precise Targeting of Previously ‘Uneditable’ Disease-related Genes
PAM stringency poses a bottleneck to therapeutic targeting of disease-associated regions that lack the preferred adjacent PAM. To test whether their relaxed and near PAMless variants could address this issue, the lab attempted to target a panel of disease-associated genomic regions that were previously inaccessible to gene editing. Remarkably, SpG and SpRY could generate biologically relevant protective genetic substitutions at single nucleotide precision in alleles associated with coronary heart disease, type 2 diabetes, and osteoporosis. The new findings potentially open up the entire human genome to gene editing therapies, raising hopes for curing genetic diseases that were thus far beyond reach.
What About Off-Target Cleavage?
When thinking about CRISPR therapies, the idea of relaxing or eliminating PAM stringency raises obvious concerns about potentially dangerous off-target effects. For instance, how can one stop a PAMless Cas9 from making edits all over the genome? While the lab doesn’t expect this first-generation PAMless Cas9 to hit the clinic any time soon, they did address off-target effects as a proof-of-concept using a method called GUIDE-seq.
Using this assay, they observed that SpG and SpRY exhibited a modest increase in off-target editing compared to wild-type SpCas9, but when known fidelity-enhancing mutations were added to the new variants, almost all detectable off-target events were eliminated. This means that it is possible to relax or remove the PAM requirement from SpCas9 without sacrificing target specificity.
CRISPR Editing and PAM Recognition in a New Light
SpG and SpRY will likely change the way scientists think about PAM recognition and CRISPR editing going forward. The most surprising aspect of the new findings is that Cas9’s PAM requirement may not be as essential as previously believed.
»The strategy behind engineering SpG and SpRY was an iterative one, whereby a variant with improved targeting capability became the basis for a further improved variant with additional modifications and so on«, explains Russell Walton. »By and large, every iteration improved the protein. We were surprised by how far we could push the apparently essential PAM requirement, yet still have a robust genome editing tool even though it can potentially interrogate any single nucleotide anywhere in the genome.«
Future Directions
SpG and SpRY, along with other Cas variants, will contribute to building better cell models or interrogating parts of the genome that were completely inaccessible before, paving the way for unprecedented functional genomics in healthy and diseased contexts. The ability to finally target the entire human genome base by base is also a welcome advance in the CRISPR field.
The lab now plans to test the new SpCas9 enzymes in various research and preclinical contexts, and efforts to engineer variants with greater PAM stringencies are also underway. As CRISPR technology evolves in research and the clinic, having a suite of Cas enzymes with different capabilities and characteristics is imperative. Imagine one day being able to pick the exact enzyme that best suits a given application, e.g., a relaxed or PAMless Cas for high-throughput functional genomics, or a very stringent Cas to cure a disease caused by a single nucleotide substitution.
Relaxed Cas9 variants such as SpG and SpRY, future variants with greater PAM stringencies, and variants with intermediate PAM requirements, will only add to the growing CRISPR-Cas9 toolbox.
The Kleinstiver Lab
The Kleinstiver Lab, headed by Assistant Professor Benjamin Kleinstiver, PhD, is located in the Center for Genomic Medicine and Department of Pathology at Massachusetts General Hospital, and the Department of Pathology at Harvard Medical School. Research in the lab is centered around three main goals: building new gene editing technologies with broad scientific applications, improving CRISPR technologies by engineering desirable properties into Cas nucleases to develop more robust, specific and safer CRISPR systems for research and therapeutics, and using these technologies to develop disease models, preclinical therapeutic strategies and understand the challenges that face the effective clinical translation of gene editing technologies.
Link to original article in Science:
Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants
Tags
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







