CasMINI: A Powerful Dwarf Among CRISPR Giants
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Groundbreaking advances have recently been made in clinical trials employing CRISPR medicines to treat patients with congenital diseases including haemoglobinopathies, ATTR amyloidosis, and Leber congenital amaurosis.
Despite this progress, the physical size of current CRISPR gene-editing nucleases limits their use in clinical applications because of the challenges involved in efficiently delivering large molecules into target cells: long coding sequences either do not fit into the cargo space of clinically safe viral delivery vectors, or they hamper the transfer capability of nanoparticles. Consequently, size is a major limiting factor for the clinical administration of gene-editing medicines.
In a hallmark paper published today in Molecular Cell, Dr. Stanley Qi and co-workers report the formation of the smallest functional CRISPR-associated protein available to date, opening doors to many new research and clinical applications.
Tailoring CRISPR with molecular engineering
Dr. Stanley Qi is an assistant professor and head of a research group in the Department of Bioengineering and the Department of Chemical and Systems Biology at Stanford University.
Over the years, his research has focused on using molecular engineering to expand gene-editing applications based on CRISPR. In 2013, he showed that by inactivating the molecular scissors of Cas9, the CRISPR nuclease can be repurposed into a programmable genetic homing device that binds specific locations in a genome without cutting the DNA. This catalytically ‘deadCas9’ (dCas9) was quickly used for a wide variety of applications, from targeted gene expression regulation (CRISPR-activation or CRISPR-interference) to visualising genomic DNA regions in live cells with CRISPR LiveFISH.
To address the challenge of delivering CRIPSR reagents into target cells, Dr. Qi and his team of scientists zoomed in on relatively compact, naturally occurring Cas nucleases as candidates for molecular engineering.
Stanley Qi explains: »The key problem we were facing is that over 99.9% of known CRISPR nucleases do not work in mammalian cells. The CRISPR defence systems evolved in prokaryotic organisms to inactivate invading phages with small and simple genomes. So, even when we solve the problem of getting the Cas nuclease into mammalian cells and across the nuclear membrane by adding localisation signals (NLS), the vast majority of natural CRISPR systems still lack potency to find and access their DNA targets among strands of millions of nucleotides, mostly tightly wrapped in chromatin.«
Given these difficult odds, Dr. Qi and his team selected a candidate from a family of very compact RNA-guided nucleases referred to as ‘class 2 type V-F’. This candidate was Cas12f , and their mission was: could the tiny Cas12f protein be made into a functional CRISPR tool for use in mammalian cells by molecular engineering?
First, the team created a robust reporter system by transforming Cas12f into a gene expression activator by neutralising the RuvC nuclease domain, and adding two NLS motifs and a strong VPR transcription activation domain. This didn't have the desired outcome though, as Dr. Qi told us: »Unfortunately, when we targeted this dCas12f-VPR to a GFP reporter system in 293 human embryonic kidney cells using previously described single guide (sg)RNAs, we observed hardly any green fluorescent cells.«
Dr. Qi and his team hypothesised that the lack of dCas12f-VPR activity could be a result of a weak interaction between the sgRNA and its target sequence.
Dr. Qi tells us how they got around that obstacle: »By making comparisons with other tracrRNAs, we generated three new RNA scaffolds to form the sgRNA. And, they all induced GFP expression in our cell system! On top of that, we engineered crucial amino acid substitutions in Cas12f that resulted in a whopping 200-fold increase in GFP expression compared to the initial version of our Cas12f-based transcription activator. We called this engineered and highly active Cas12f version ‘CasMINI’.«
“In this paper, we present the first proof-of-principle that molecular engineering of a non-functional Cas protein can result a in working, programmable, multifunctional CRISPR tool.”
A multifunctional miniature Cas protein
Follow up experiments quickly showed that the compact CasMINI functioned as well as much bigger and established CRISPR proteins like Cas12a (Cpf1). When targeted to the promoter regions of endogenous genes, dCas12f-VPR induced equal or even better expression levels when compared to a highly similar CRISPR transcriptional activator based on Cas12a. As far as off-target gene activation goes, no outliers were identified based on whole transcriptome analysis, on par with the highly specific Cas12a. »This showed that CasMINI is highly specific and we haven’t detected off-target effects,« emphasises Dr. Qi.
To determine whether the functional capabilities of CasMINI could be extended, a fusion protein was made with a base editor, dCasMINI-ABE, to create A-T to G-C nucleotide conversions. This worked out very well, as Dr. Qi explains: »It turned out that dCasMINI-ABE is effective in making A-T to G-C DNA changes, with peak base-editing activity occurring at a narrow window 3 to 4 base pairs downstream of the PAM«, the protospacer adjacent motif ‘TTTR’ just upstream of the crRNA binding region.
And did the engineered sgRNA and protein modifications create programmable DNA scissors that could cut DNA? »Yes, indeed!« answers Dr. Qi.
»When we duplicated our amino acid changes in Cas12f with an active RuvC domain, we observed target deletions in the genomic sequences. This indicated that the amino acid changes that optimised expression also promoted nuclease activity. However, we saw that the CasMINI optimised for expression activation was somewhat less active in creating double-strand DNA breaks than an earlier Cas12f variant with fewer amino acid substitutions in the vicinity of the RuvC domain.« elaborates Dr. Qi.
Nuclease-active CasMINI and derived variants create a DNA break just downstream of the crRNA binding site. The observed deletions are much larger in comparison to those derived from standard Cas9 activity, amounting to 20 nucleotides on average. These bigger deletions imply that CasMINI can likely be more effective than Cas9 for fully disrupting gene function.
CasMINI makes a difference
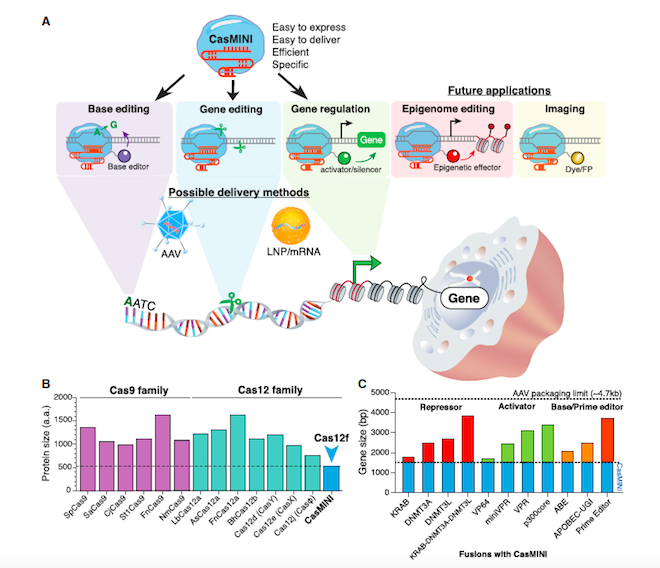
In the Molecular Cell paper, first author Dr. Xiaoshu Xu and colleagues present the first proof-of-principle that molecular engineering of a non-functional Cas protein can result a in working, programmable, multifunctional CRISPR tool (Figure 1A).
»Obviously, there are still many things to optimise or validate,« says Dr. Qi. »For instance, we can see base-editing bystander effects, where additional A-T nucleotides in the vicinity of the target may also be converted into G-C base pairs. However, other studies have already shown ways to enhance the precision of base editors. Furthermore, we would like to do more experiments to carefully assess the specificity of the nuclease active CasMini.«
So how much smaller is CasMINI compared to other Cas proteins? »The engineered CasMINI protein consists of 529 amino acids. This is about 2.5 times smaller than other frequently used CRISPR proteins« like Cas9 from Streptococcus pyogenes or Cas12a from Lachnospiraceae bacterium explains first author, Dr. Xiaoshu Xu (Figure 1B).
Previously, other small CRISPR proteins like CasX or Cas-Phi have been reported, but these are still approximately 2 or 1.5 times larger than CasMINI, respectively. These differences are clinically significant, as the coding sequence of CasMINI can easily be accommodated in important in vivo delivery tools like adeno-associated virus (AAV) platforms. The cargo space in AAV is about 4.7 kb, this means that even CasMINI fusion constructs with transcription regulators, base editors, or potentially PRIME editors will fit within AAV viral particles. »There is even room left for an additional sgRNA expression cassette!« adds Dr. Xu (Figure 1C).
A big clinical future?
Thanks to its tiny size and functional features, CasMINI has really favourable characteristics for clinical development, and this is what Stanley Qi and the team are aiming for: »The delivery advantages of CasMINI are clear. But the small size may also reduce immune reactivity in patients – there are fewer epitopes compared to established CRISPR enzymes. Also, Cas12f is derived from a species that doesn’t use humans as a host. That’s very important. Patients won’t have pre-existing antibodies against CasMINI like they do for spCas9. This might boost therapy efficiency and avoid immune response-associated complications.«
Asked about which inherited medical conditions to aim for first, Dr. Qi points to recent clinical trials as a starting point. »But, there are many unmet medical needs that cannot be tackled with current CRISPR medicines. CasMINI could be really important for expanding the clinical applications of CRISPR. It could become a big player in CRISPR medicine.«
CasMINI and variants described in the publication will soon be available to the scientific community via Addgene.
Link to original article in Molecular Cell:
Engineered Miniature CRISPR-Cas System for Mammalian Genome Regulation and Editing.
Henri van de Vrugt is a molecular biologist and Chief Scientific Officer at New Haven Biosciences Consulting, US.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







