CAST-Seq: A New and Much-Needed Tool for Assessing the Safety of Designer Nucleases
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Before any therapy based on gene-editing technology can be deemed safe, the specificity and activity of the nuclease in question, i.e., the Cas, TALEN, zinc finger nuclease (ZFN) or otherwise must be thoroughly evaluated to ensure that the integrity of genome-edited cells is maintained and to minimise the risk of oncogenic transformation following therapeutic administration.
Toni Cathomen is a Professor and Director of the Institute for Transfusion Medicine and Gene Therapy at the University of Freiburg, where the development of new therapies based on gene editing has been a long-standing focus.
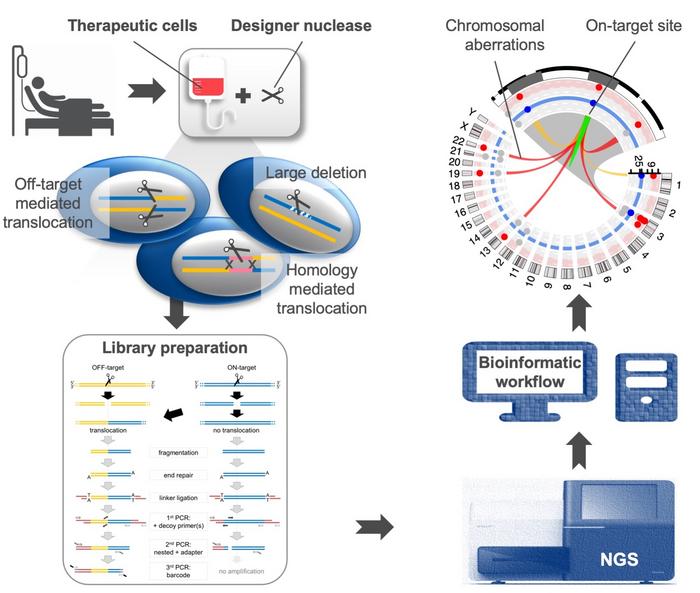
Cathomen’s group recently developed a new tool called ‘chromosomal aberrations analysis by single targeted linker-mediated PCR sequencing’ or CAST-Seq that can identify and quantify chromosomal rearrangements following gene editing with CRISPR-Cas and TALENs, marking a much-needed addition to the toolbox for assessing the safety of designer nucleases. The group's findings were published recently in Cell Stem Cell.
Off-target assays often don’t reveal the full picture
»The existing assays tend to focus on small off-target insertions and deletions (indels) and not the larger chromosomal rearrangements that may be induced by gene-editing nucleases. It has been known for a while that if there is on- and off-target activity at the same time, chromosomal translocations can occur. We believe it is important to get the full picture of what happens in a cell and not just the off-target edits, but also the full effect of these. So we developed CAST-Seq as a preclinical assay that could do exactly that,« explains Toni Cathomen about the motivation behind developing CAST-Seq.
CAST-Seq is an assay that can detect and quantify gene-editing induced chromosomal aberrations with high sensitivity by mapping the chromosomal sequences fused to one arm of the target site. In other words, if a translocation has occurred at the target site, the assay can reveal what chromosome/chromosomal region the target site is now fused to.
The group has spent three years developing the CAST-Seq in vitro and bioinformatics workflow and Toni Cathomen is excited about the outcome:
»We found novel kinds of chromosomal rearrangements that have never been described before. Our assay makes it feasible to quantify all of these chromosomal rearrangements in therapeutically relevant gene-edited cells for the first time.«
The CAST-Seq workflow
The in vitro part of the CAST-Seq workflow is based on a three-step PCR reaction.
The workflow begins with isolation and fragmentation of genomic DNA from gene-edited cells. Linkers are added to these DNA fragments, which allow amplification and identification of regions of interest later on. Four primers are then added. The first is an anchor or ‘bait’ primer that binds to the target sequence on one side of the double-stranded break (DSB) that was introduced with CRISPR-Cas or a TALEN. The second primer is a ‘prey’ primer that binds to the linker sequence.
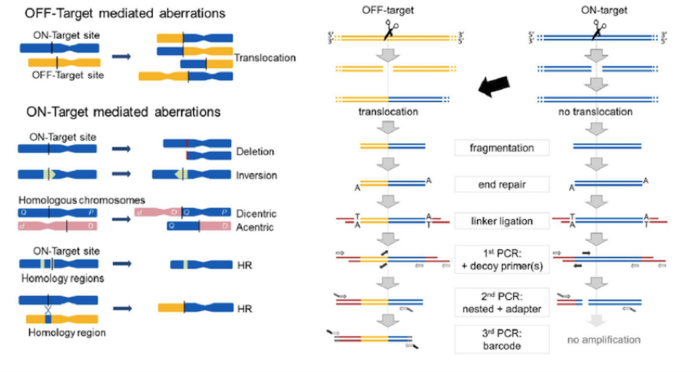
To circumvent the fact that translocations are very rare and to reduce background noise so as to detect rare and unusual events, the group came up with a smart trick to ‘get rid’ of DNA fragments that don’t contain a translocation. This involved the addition of a set of nested locus-specific ‘decoy’ primers that bind to the target region within the amplicon that would be generated by the bait and prey primers.
In the absence of any translocation the decoy primers prevent amplification with the bait and prey, but a translocation or large deletion at the target site precludes binding of the decoy primer, thus allowing amplification with the bait and prey primer pair. The inclusion of decoy primers was an innovative move that allowed Cathomen to patent the CAST-Seq methodology, and a company start-up to further progress the technology is around the corner.
The resulting amplicons from the CAST-Seq reaction are then sequenced and analysed with a purpose-built bioinformatics pipeline that can classify the chromosomal aberrations into various categories of on- or off-target aberrations.
CAST-Seq reveals on-target homology-mediated translocations
Once the assay was up and running, the group performed CAST-Seq in primary human haematopoeitic stem and progenitor cells (HSPCs) following gene editing at various time-points. They selected a panel of target sites that were edited by either wild-type Cas9 or a high-fidelity Cas9 variant, each delivered as ribonucleoprotein (RNP) complexes, or a TALEN delivered in mRNA format.
On-target activity was determined to be in the range of 48-89 % measured by indel formation, with cell viabilities ranging from 70-99 %. CAST-Seq detected translocations in 0-0.5 % of gene-edited HSPCs, and up to a fifth of on-target loci harboured gross chromosomal rearrangements.
Bioinformatic analysis of the translocations picked up by CAST-Seq revealed not only translocations at off-target sites as expected, but also chromosomal rearrangements at on-target sites. This was a surprise, as Toni Cathomen explains:
»We also found translocations that we knew could not be the result of off-target activity, because we could not find any off-target sites. What we did find within these translocations was a stretch of at least 25 nucleotides long with homology to the on target site. So, if you have a genomic sequence that is similar in sequence to your target site, you don't need a second break, but because the chromosome is now broken, it looks for something like the sister chromatid. If it finds homologous sequence somewhere else in the genome, it will take it to repair the DSB. We believe this to be how these homology-mediated translocations can occur, and we could validate the nature of these translocations by direct PCR.«
A serendipitous discovery
Cathomen also shared that the discovery of homology-mediated translocations was an act of serendipity. The first target that the group looked at was the CCR5 locus, and right next to this is the CCR2 gene. These two genes probably evolved from a gene duplication event, as they’re very similar. A lot of the large deletions uncovered by CAST-Seq were in fact huge rearrangements spanning 13 or 14 KB of DNA, but in the case of CCR5 and CCR2, the group could not find a second DNA break because CCR2 was not cleaved by the nuclease – they could show this.
»Finding homology-mediated translocations was actually a massive surprise because no one described it before and we could only do it because our assay is so sensitive. To test our hypothesis that this was down to homology, we targeted the HBB locus, which is right next to the HBD locus. These haemoglobin genes are also the result of an earlier gene duplication, and we got a similar result here – homology-mediated translocation,« said Cathomen.
Where does CAST-Seq fit into the therapeutic gene editing process?
Toni Cathomen sees CAST-Seq as a novel diagnostic tool that can be used to quality control gene-edited cells, e.g., blood stem cells, before they are transplanted into a patient. This potential makes CAST-Seq distinct from other off-target assays out there, as Cathomen explains:
»CAST-Seq is not a predictive assay as most of the available assays are. The typical off-target assays are performed in vitro. These can tell us where the potential off-target sites are, and one can then go back and check in the cell of interest whether or not these sites were cleaved. But with CAST-Seq, we are looking directly in the cells that will be transplanted to the patient.«
Early on in the development of a new therapeutic, CAST-Seq may be applied to help identify nucleases, i.e., Cas, ZFN, and TALENs, whose specificity profiles warrant progression to clinical development. The group is already collaborating with a number of companies in the gene-editing space to explore the potential of CAST-Seq in the lead selection process.
Returning to the diagnostic application, Cathomen explains how CAST-Seq could hypothetically fit into a clinical setting for approved therapies (hypothetical because there are as-of-yet no approved gene-edited therapies):
»A clinical site can apply the assay before gene-edited cells are transplanted to a patient, and this could become a part of the release criteria for every single gene-edited cell product. Current quality control procedures for stem cell transplants take about two weeks, which leaves plenty of room to run CAST-Seq on a portion of the cells in the meantime. One could then confidently say that there are no translocations within the assays limit of detection, and this could be used to set fixed safety limits for the product.«
Cathomen then describes how CAST-Seq may also be a useful safety measure in ongoing clinical trials to monitor the development of gene-edited stem cells over time. Bone marrow biopsies are very painful and no one wants to do this to a patient unless absolutely necessary. Instead, one can look at peripheral blood cells since these are derived from the stem cells.
»Using CAST-Seq, it is possible to detect clonal expansion and examine clonal dynamics within patients as an early indication of safety. If one clone suddenly becomes dominant, this may be a sign that something is going wrong in this patient. One can then closely monitor the patient to see whether she/he develops a leukaemia,« explains Cathomen.
Future Directions
On a research level, CAST-Seq has the potential to reveal insights into the DNA repair mechanisms at play during gene-editing events and the group is currently exploring how the nuclease delivery format may impact the outcome as well as how CAST-Seq can be used to monitor the activity of other gene-editing modalities that are edging towards the clinic, such as CRISPR base editors.
The group observed that the total number of translocations reduced over time following genome editing by Cas9 or TALEN. In most cases, the nucleases were active for about 24 hours and the formation of translocations were detected for approximately 48 hours after which time some translocations were lost. Further experiments will help to unravel whether these ‘lost’ translocations were associated with certain deleterious states such as acentric (no telomere) or dicentric (two telomeres) chromosomes that killed the gene-edited cells thus eliminating them from the analysis.
From a therapeutic point of view, translocations that kill gene-edited cells before they are transplanted into a patient might not be a major worry, but translocations in general need do to be taken into account during development and alongside off-target quality control of gene-edited therapies, as Toni Cathomen points out:
»What we have to be concerned about is translocations that can link a very strong promoter to a proto-oncogene and trigger leukaemia. CAST-Seq can be used to make a clinical risk assessment on gene-edited cells based on the translocations that are found.«
Link to original article in Cell Stem Cell:
Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq
Tags
ArticleInterviewNewsin vivoRibonucleoprotein (RNP)AI in genome editingGene therapyOff-targetQuality ControlCRISPR-CasTALENs
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







