Conditional CRISPR Offers Logical Regulation of Gene Networks
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
A significant challenge in synthetic biology is the regulation of gene networks rather than single genes. Traditional methods often struggle to control multiple genes simultaneously due to the complexity and interconnectivity of gene networks. This complexity can result in unpredictable behaviours and reduced efficiency in gene expression control. The precision required to manage multiple genes, especially within metabolic pathways, demands sophisticated tools that can integrate various signals and respond accurately, something that has proven difficult with existing technologies.
The concept of logical operations, such as those found in digital electronics, offers a promising solution for the regulation of gene expression. By employing Boolean logic gates (AND, OR, NOT, and their combinations), researchers can create genetic circuits that perform complex decision-making processes based on multiple inputs. Logic gates allow for the conditional regulation of genes, providing a more robust and flexible approach to controlling biological systems. Logical operations enable the integration of diverse signals, facilitating precise control over gene networks and enhancing the ability to fine-tune cellular processes.
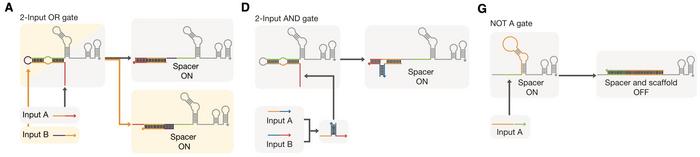
In this study, the researchers combined a dCas9-effector (fused with a transcription activating protein) with conditional gRNAs (cgRNAs) that operate by incorporating a trigger RNA that activates them through a specific interaction (Figure 1). In the absence of the trigger RNA, the cgRNA remains inactive, preventing gene expression. When the trigger RNA is present, it binds to the cgRNA, inducing a conformational change that exposes the spacer sequence. This allows the cgRNA to guide the CRISPR-Cas complex to the target gene for regulation. This mechanism enables precise control of gene expression in response to specific molecular signals.
The cgRNAs were designed with an optimised secondary RNA structure and molecular ratios to achieve tight control over gene expression. This system allowed for the construction of various Boolean logic gates, enabling the integration of multiple inputs to regulate target genes. The cgRNAs demonstrated a high dynamic range and low leakage, making them practical tools for precise genetic regulation even in complex biological circuits (Figure 2).
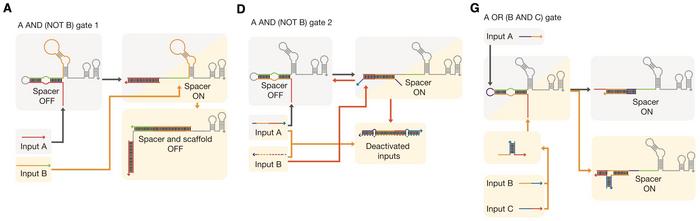
The team applied these cgRNAs to regulate essential E. coli genes involved in metabolic processes and cell division, such as lacZ, malT, poxB, ftsZ, and mreB. By combining different logical gates, the researchers were able to create sophisticated genetic circuits that could process complex inputs and control gene expression predictably. The study showcased the ability to control cellular morphology and metabolic flux, highlighting the potential of cgRNAs in various applications, from metabolic engineering to synthetic biology.
The findings of this study provide a new perspective on the regulation of gene expression using programmable gRNAs, with significant implications for human clinical applications. The ability to implement logical operations within genetic circuits involved in complex diseases enhances our capability to design gene therapies that can make sophisticated decisions based on multiple biological signals. Thereby, the novel study potentially paves the way for more effective and personalised treatments for conditions such as cancer, genetic disorders, and metabolic diseases.
Hansol Kang, Dongwon Park, and Jongmin Kim at Pohang University of Science and Technology, Korea, led the study, which was published in Nucleic Acids Research on 29 June 2024.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.