CRISPECTOR2.0 Quantifies Allele-Specific Genome Editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The human genome contains numerous single-nucleotide variants (SNVs), which contribute to differences between gene copies and among individuals. SNVs at on-target or off-target sites can either enhance or reduce Cas9 activity by modifying the nuclease recognition sequence. However, existing pipelines, including CRISPECTOR (previously featured in CRISPR Medicine News, see below), typically rely on a single reference genome for CRISPR activity analysis or require prior knowledge of the variants (alleles) in the sample.
CRISPECTOR2.0 improves upon this by incorporating an SNV detection feature that identifies SNVs specific to each sample, creating a 'personalised reference genome' that reflects the individual's alleles. It then quantifies insertions and deletions (InDels) induced by Cas9 for each allele independently, providing a precise, allele-specific profile of Cas9 activity (see Figure 1).
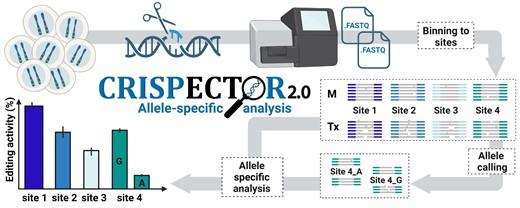
»We screened a dataset of 40 gRNAs and identified a few interesting cases of 'differential editing activity', in which the CRISPR editing activity was limited to only one allele. This highlights the importance of analysing CRISPR-Cas9 editing activity in an allele-specific manner,« says Nechama Kalter. She is a PhD student at Bar-Ilan University, Israel, and co-first author of a paper that was recently published in Nucleic Acids Research (1).
Nechama Kalter developed the new tool together with the paper's other co-first author, Guy Assa, a machine learning and data scientist at Reichman University. By applying a statistical algorithm, CRISPECTOR2.0 calls alleles based on sequencing data and calculates genome-editing frequencies for each allele. This method was validated across diverse cell types, including human cells and plants, highlighting significant editing variations even in regions with distant SNVs. These findings underline the importance of measuring allele-specific editing for safer, more efficient genome-editing applications.
“CRISPECTOR2.0 significantly advances the field of genome editing for therapeutic purposes by offering personalised on-target and off-target evaluations, without the need for prior knowledge about the individual's specific genome or any additional experiments”Guy Assa
»CRISPECTOR2.0 significantly advances the field of genome editing for therapeutic purposes by offering personalised on-target and off-target evaluations, without the need for prior knowledge about the individual's specific genome or any additional experiments. This tool provides a robust and tailored assessment, ensuring that each patient's data is accurately evaluated for both safety and efficiency,« Guy Assa explains.
CRISPECTOR2.0 is designed to be user-friendly and simple to use. The installation is straightforward via Bioconda or Pip platforms, with detailed instructions provided on CRISPECTOR2.0's GitHub page. Once installed, its usage mirrors that of CRISPECTOR: a single command line input generates a comprehensive HTML file, detailing the editing activity at both on- and off-target sites, including allele-specific information.
The research was led by Ayal Hendel and Zohar Yakhini, who are affiliated with Bar-Ilan University and Reichman University, respectively. It was published on 30th July 2024 in Nucleic Acids Research
1) Assa et al. (2024). Quantifying allele-specific CRISPR editing activity with CRISPECTOR2.0. Nucleic Acids Research, https://doi.org/10.1093/nar/gkae651
2) Palmgren (2021). CRISPECTOR Accurately Detects Translocations and Off-Target Activity. CRISPR Medicine News, https://crisprmedicinenews.com/news/crispector-accurately-detects-translocations-and-off-target-activity/
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







