CRISPR Helps Decode the Epigenetic Mysteries of Cancer
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Since the early days of her academic journey, Gabriella Ficz has been deeply engaged in epigenetics. She initially focused on polycomb group proteins in Drosophila before pivoting to mammalian DNA methylation. Despite that shift, her long-term interest in epigenetics has always centred around its crucial role in maintaining cell identity.
Currently a group leader at Queen Mary University of London, she is now committed to dissecting the epigenetic alterations that underlie the onset of cancer.
One of the challenges in studying epigenetics is the issue of correlation versus causation. Multiple changes co-occur, making it difficult to pinpoint what drives what.
To disentangle these complexities, Gabriella Ficz has turned to CRISPR technology, and last month, her most recent findings were published in an article in PNAS with postdoctoral research assistant fellow Emily Saunderson as the first author. Emily Saunderson obtained a prestigious Kay Kendall junior fellowship to undertake this work.
The team used catalytically inactive Cas9 (dCas9) fused to a DNA methyltransferase to alter only one specific epigenetic locus in perfectly healthy cells without changing the genetic makeup, allowing them to see the effects of that aberrant DNA methylation.
Gabriella Ficz explains:
“Our main objective was to understand the epigenetic signatures observed in AML. Although AML pathogenesis is complex, involving genetic and epigenetic alterations, many AML cases exhibit these aberrant epigenetic patterns, even when different sets of genes are mutated”Gabriella Ficz
» We wanted to keep the study design as clean as possible by focusing on one genomic locus rather than many. Epigenetic signatures usually involve multiple targets across the genome, which can introduce numerous variables and complicate the interpretation of results. By narrowing our focus, we aimed to get as close to black-and-white answers as possible, to reduce the complexity and better disentangle causation from correlation.«
CRISPR epigenetic editing targets p15
Previous research has shown that different subtypes of acute myeloid leukaemia (AML) exhibit a particular epigenetic signature at the promoter of the tumour suppressor gene p15. When the team consolidated data from multiple studies, 60% to 80% of AML cases appeared to have this epigenetic pattern. The observation led them to consider whether these changes occur coincidentally in all instances or perhaps emerge early in the pathogenesis of the disease.
» Our main objective was to understand the epigenetic signatures observed in AML. Although AML pathogenesis is complex, involving genetic and epigenetic alterations, many AML cases exhibit these aberrant epigenetic patterns, even when different sets of genes are mutated. We wanted to know if these epigenetic changes are pathogenic in the same way mutations are,« says the epigeneticist.
Gabriella Ficz and her team used CRISPR epigenetic editing to target hypermethylation of the p15 promoter in CD34+ primary human haematopoietic stem and progenitor cells (HSPCs) obtained from cord blood. The CRISPR tools were delivered using nucleofection, i.e., electroporation with ribonucleoprotein (RNP) complexes of the dCas9-effector fusion protein and synthetic guide RNAs.
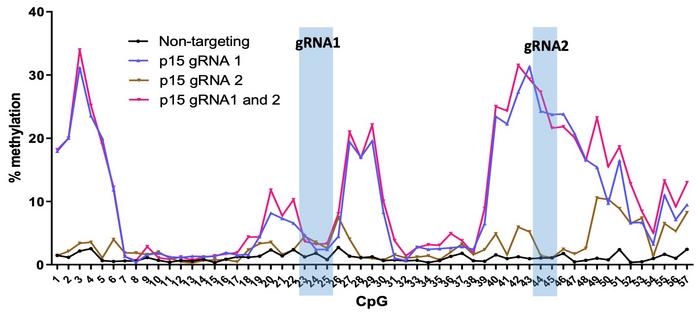
Two weeks after nucleofection, bisulphite sequencing revealed that targeted DNA methylation spread across 300-500 base pairs of the p15 promoter with up to 43% methylation at individual CpGs (see Figure 1). In contrast, controls with non-targeting gRNA were virtually unmethylated, and genome-wide analyses showed minimal to no off-target DNA methylation events.
The DNA methylation pattern across the p15 promoter was highly reproducible across different experiments but depended on the gRNA position. This was shown by very little methylation obtained with a second gRNA targeting a location further downstream of the CpG island, even though the first gRNA heavily methylated this location.
Hypermethylation is maintained during differentiation
Interestingly, hypermethylation of the p15 promoter was maintained when the epigenome-edited haematopoietic stem cells differentiated into myeloid and lymphoid cell lineages. Maintenance of methylation was the case both in vitro and in vivo when edited stem cells were engrafted into the bone marrow of immunodeficient mice (see Figure 2).
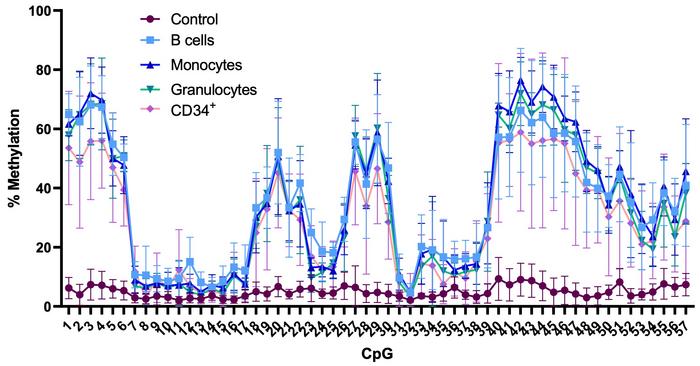
»We were very surprised that the DNA methylation pattern is so heritable in the in vivo system. Generally, when people edit DNA methylation in cell lines, the methylation pattern is not faithfully inherited, or at least not for long. We still don't fully understand why primary cells seem more receptive to heritable methylation,« says Gabriella Ficz.
“The data suggest that these epigenetic changes induce an inflammatory environment, which we hadn't anticipated. This could potentially increase cell proliferation, leading to more mutations and contributing to cancer”Gabriella Ficz
Differentiated cells with hypermethylation at p15 consistently had almost 50% lower expression of this gene. Moreover, transcriptome analysis revealed that engraftment of p15 hypermethylated HSPCs into mice gives rise to monocytes and granulocytes with distinct immune effector functions.
Compared to controls, monocytes showed up-regulation of genes involved in antigen presentation (HLA genes) and proinflammatory cytokines (IL1B, TNF) (see Figure 3F). In granulocytes, receptors (DICER1, TLR2) and transcription factors involved in pathogen recognition and the type I interferon response (JUN, NFAT5) were differentially expressed (see Figure 3G).
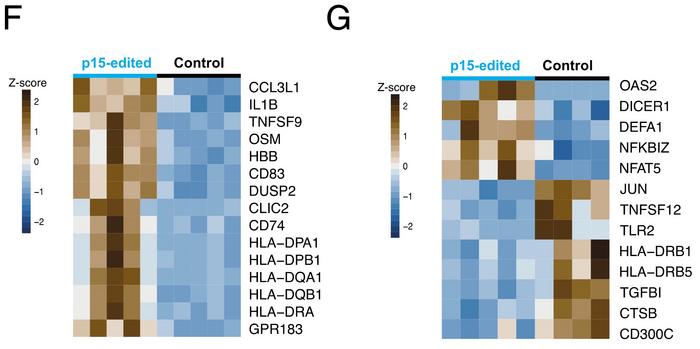
Gabriella Ficz was surprised by the immune phenotype she observed:
»The data suggest that these epigenetic changes induce an inflammatory environment, which we hadn't anticipated. This could potentially increase cell proliferation, leading to more mutations and contributing to cancer. But it is still a hypothesis that needs to be tested.«
DNA methylation of p15 may be clinically relevant
Though the study indicates that hypermethylation of p15 could have adverse effects, she doubts that reversing the process in cancer cells – e.g., by using CRISPR to demethylate p15 – can have a potential role in cancer therapy:
» It's inaccurate to say that hypermethylation of p15 directly induces cancer. We know that hypermethylation of p15 impacts haematopoietic stem cell differentiation and affects immune cells. As for its potential to trigger cancer, we haven't conducted long-term studies yet. So, the idea about manipulating methylation levels in cancer cells is exciting and worth exploring, but given the complexities of cancer, I'm uncertain if it would be an effective treatment.«
“I think cancer cells could potentially circumvent any epigenetic interventions unless we target numerous genomic locations. However, I can speculate that it might be feasible to use epigenetic editing to manipulate immune cells to target cancer cells”Gabriella Ficz
Still, DNA methylation of p15 may be clinically relevant. In the PNAS paper, Gabriella Ficz refers to previous research that has quantitatively assessed p15 promoter hypermethylation in the bone marrow of healthy subjects compared to patients with progressive stages of myeloid disease. That study found 35 CpGs where methylation increases as the disease progresses, and 54% of those coincide with sites with greater than 10% hypermethylation in Gabriella Ficz's analysis.
To clarify if p15 hypermethylation could be involved in the early stages of AML, the London-based epigeneticist also compared methylation of p15 between healthy controls and patients with clonal haematopoiesis of indeterminate potential (CHIP), which is a non-malignant condition that may progress to AML in some patients. Though no significant differences in the mean methylation level were found between the two groups, the data indicated that p15 hypermethylation is detectable in the peripheral blood of some patients with CHIP, and pre-AML patients have increased variability of methylation at this promoter.
»When it comes to translating these findings into clinical applications, there are two aspects to consider. The first aspect is understand the early stages of the disease, which we've been focusing on. Secondly, we're interested in using CRISPR tools, particularly for epigenetic editing, to influence cells within the tumour microenvironment or the tumour cells themselves,« says Gabriella Ficz and continues:
» I'm not yet convinced that directly altering the epigenetic state of cancerous cells would be effective, primarily due to the strong evolutionary forces at play. I think cancer cells could potentially circumvent any epigenetic interventions unless we target numerous genomic locations. However, I can speculate that it might be feasible to use epigenetic editing to manipulate immune cells to target cancer cells.«
Link to the original article in PNAS:
CRISPR/dCas9 DNA methylation editing is heritable during human hematopoiesis and shapes immune progeny (2023) Proc. Natl. Acad. Sci. USA 120: e2300224120
For more information, see:
CRISPR/Cas9-Targeted De Novo DNA Methylation Is Maintained and Impacts the Colony Forming Potential of Human Hematopoietic CD34+ Cells (2019) Blood 134: 2517
A gentler way to tweak genes: epigenome editing (2022) Science 376: 1034
New insights into mechanisms that regulate DNA methylation patterning (2015) The Journal of Experimental Biology 218: 14
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.

The First CRISPR Medicine Conference, Copenhagen April 2024 - More details soon...
Tags
ArticleInterviewNewsElectroporationAcute Myeloid Leukemia, AMLCancerEpigenome editing (e-GE)dCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.