CRISPR Identifies New Genes as Drivers of Prostate Cancer Metastasis
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Prostate cancer is the second most common cancer worldwide, claiming around 375,000 deaths in 2020. It often progresses to a metastatic stage, where cancer cells spread beyond the prostate gland to other parts of the body, making treatment more complex and reducing survival rates. Traditionally, the focus of prostate cancer research has been on early detection and localised treatment. However, the challenge of preventing and managing metastasis has remained a formidable barrier.
The recent identification of three novel metastasis-driving genes opens up new possibilities for intervention at a molecular level. By targeting these specific genes, researchers ultimately hope to develop more effective treatments that can halt the spread of cancer cells, improve patient outcomes, and transform the prognosis for those affected by advanced prostate cancer.
»In prostate cancer and many other solid tumours, metastasis is the ultimate cause of death. Therefore, there's a pressing clinical need to identify the drivers of metastasis and understand the molecular basis behind the transition from indolent to aggressive tumours,« says Juan Martín Arriaga, who is an assistant professor at Icahn School of Medicine at Mount Sinai, New York. He is the first author of a paper published earlier this year in Oncogene that describes the use of an in vivo genome-wide CRISPR screen, identifying CITED2 as a novel driver of prostate cancer bone metastasis.
Epigenetic factors are crucial for metastasis
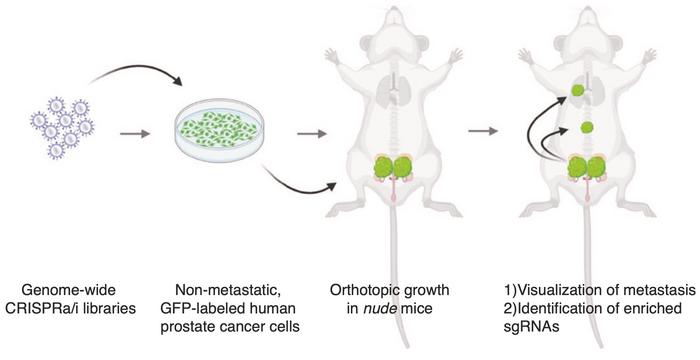
In his experimental approach, Juan Martín Arriaga injected human prostate cancer cells into immunodeficient mice (see Figure 1). Without any modification, there was no evidence of metastasis outside the prostate or lymph nodes, which made this a good model. He explains:
»We aimed to conduct unbiased screens to query all genes at the genome-wide level. Using CRISPR activation and inhibition, we activated or inhibited genes one at a time to identify any that could drive metastasis. If activating a gene led to metastasis in a previously non-metastatic tumour, we defined it as a metastatic driver. Conversely, inhibiting a gene that allowed metastasis to occur defined it as a metastasis suppressor. Our results show that if we overexpress CITED2 in the prostate, we see metastasis.«
“In tumours without Kmt2c mutations, we don't see metastases. This tells us that KMT2C is essential - it's a smoking gun”Martin Kristian Thomsen
Martin Kristian Thomsen, an assistant professor at Aarhus University in Denmark, took another approach in his study, which was published in Nature Communications on the same day as Juan Martín Arriaga's. He used CRISPR to specifically mutate five tumour suppressor genes (Pten, Trp53, Rb1, Stk11, and RnaseL) and three epigenetic factors (Kmt2c, Kmt2d, and Zbtb16) in the mouse prostate.
The team developed a Cas9-positive mouse model with prostate-specific expression, allowing targeted mutations and preventing off-target tumours (see Figure 2). With this setup, screens could be carried out by combining up to all eight sgRNAs in a single adeno-associated virus (AAV) and injecting it into the mouse prostate. Targeting all eight genes at once, however, yielded mice with very complex phenotypes. So, they had to separate the epigenetic factors.
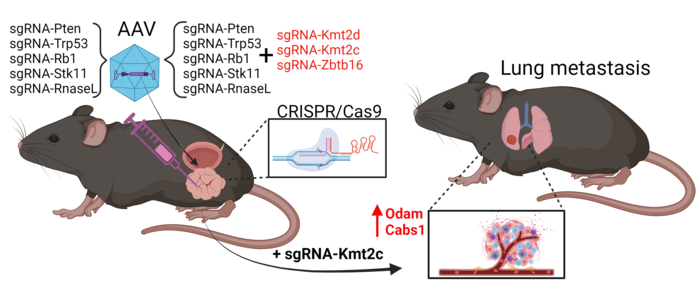
»We hypothesised that some genes are crucial for initiating cancer, while others are important later. When we target the five suppressor genes, we can see tumours proliferate, but no metastasis occurs. Next, we target these five genes and also combinations of the epigenetic factors ZBT616, KMT2D, and KMT2C. When scanning the lungs, we now see metastases,« says Martin Kristian Thomsen.
Detailed analysis revealed that Kmt2c is crucial for metastasis. When KMT2C was mutated, it consistently led to metastases in the lungs, whereas other factors like ZBT616 and KMT2D did not have the same effect. This finding was confirmed through whole genome sequencing and genotyping, highlighting the specific role of KMT2C in cancer spread.
»In tumours without Kmt2c mutations, we don't see metastases. This tells us that KMT2C is essential - it's a smoking gun,« Martin Kristian Thomsen states.
Complex regulatory networks define cancer progression
But CITED2 and KMT2C are not the only drivers of prostate cancer metastasis. Just a few days before the two papers mentioned above were published, an article in Cancer Letters revealed that PRMT7 is also essential for metastasis. First author Maria Rodrigo-Faus and corresponding author Alvaro Gutierrez-Uzquiza, both from Complutense University of Madrid in Spain, led the study.
The team conducted high-throughput CRISPR-Cas9 screens to identify genes essential for the metastatic process. The screens were performed on Cas9-positive human prostate cancer cell lines DU145 and PC3 – both highly metastatic - using a gRNA library targeting 20,000 genes. Subsequently, an in vitro invasion assay was conducted to identify genes essential to the invasive process (see Figure 3).
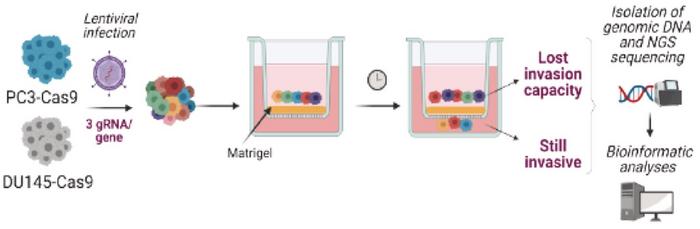
Among numerous genes whose depletion significantly impaired the cells' invasive abilities, PRMT7 emerged as a significant candidate. Further validation using siRNA technology confirmed that inhibiting PRMT7 significantly reduced the invasive capacity of the two cell lines. A similar reduction of metastatic dissemination was observed after CRISPR-Cas9 depletion of PRMT7 in in vivo assays, including chicken embryo and mouse xenograft models.
The three teams also investigated how the genes identified as crucial in driving prostate cancer metastasis operate on the molecular level. One of the genes, CITED2, is a transcriptional regulator and was shown to interact complexly with the transcription factor E2F1.
»We found that when CITED2 upregulates E2F1, which is associated with proliferation and metastasis, those tumours become metastatic. In some tumours, CITED2 does the opposite; it downregulates E2F1. We think that, initially, CITED2 inhibits E2F1 expression in primary tumours, preventing metastasis. However, as the tumour evolves or in specific subtypes of cells, CITED2 can activate E2F1, driving metastasis. It's not just the overall expression level of CITED2 but its interaction with E2F1 that determines the metastatic outcome,« explains Juan Martín Arriaga.
To delve deeper into these findings, the American team is planning follow-up studies, focusing on specific pathways or gene groups to reduce variability and enhance the accuracy of results. Additionally, the team collaborates with Columbia University to use a multi-organ bioreactor, or “organ-on-a-chip”, to mimic the metastatic environment. This bioreactor allows cells to circulate and potentially invade engineered tissues like bone, providing a sophisticated invasion assay to study metastasis in a controlled setting.
The three genes have therapeutic potential
CITED2 itself is not directly druggable because of these complex interactions and because we don't have small molecules that can selectively inhibit its activity. However, its role as a transcriptional regulator suggests that targeting its interactions with specific transcription factors could be a viable strategy.
»If we could find small molecules that selectively inhibit its binding to the partners driving metastasis, then it could be druggable,« Juan Martín Arriaga suggests.
The molecular mechanisms of KMT2C and PRMT7 are different from those of CITED2 and might have more promising therapeutic potential. PRMT7 catalyses arginine methylation primarily in histones. It methylates both activating and repressive histone marks, thereby contributing to the epigenetic regulation of gene expression.
“When we used CRISPR to knock out Odam and Cabs1, no metastases were observed in the fat, lymph nodes, or lungs. This suggests these genes are important for metastasis, regulated by KMT2C knockout”Martin Kristian Thomsen
Higher expression levels of PRMT7 are known to correlate with increased tumour aggressiveness and poor survival rates in prostate cancer patients. Maria Rodrigo-Faus and her team also showed that pharmacological inhibition of PRMT7 using available inhibitors significantly reduced the invasive capabilities of prostate cancer cells in vitro. This suggests that targeting PRMT7 could be a promising therapeutic strategy for managing metastatic prostate cancer.
KMT2C likewise encodes a histone methyltransferase that regulates gene expression through modification of chromatin structure. Such epigenetic regulation is crucial because it opens and closes the chromosome, allowing transcription to occur.
»We looked for global changes in epigenetic regulation but couldn't pinpoint any. However, through RNA sequencing, we found a cluster on chromosome 5 in the mouse, analogous to a cluster on human chromosome 4, where certain genes were highly expressed in metastases,« Martin Kristian Thomsen recalls.
His team demonstrated that loss of KMT2C increases gene expression in this 150 kb conserved region, which includes the two genes Odam and Cabs1. These two genes were highly expressed in metastases, suggesting a crucial role in cancer progression. The Danish researcher explains:
»When we used CRISPR to knock out Odam and Cabs1, no metastases were observed in the fat, lymph nodes, or lungs. This suggests these genes are important for metastasis, regulated by KMT2C knockout.«
Martin Kristian Thomsen highlights the potential for integrating mutation analysis into clinical practice. By sequencing patient tumours to detect KMT2C mutations, clinicians could identify those at higher risk for metastasis, enabling targeted surveillance and personalised treatment plans. This approach could improve outcomes, especially as sequencing becomes more affordable and accessible, offering a more precise and cost-effective way to manage cancer treatment.
Collectively, the three studies underscore the potential of targeting PRMT7, CITED2, and KMT2C, including their transcriptional targets, as therapeutic strategies to mitigate metastasis and improve outcomes in prostate cancer patients. Moreover, the genes might potentially serve as biomarkers for monitoring prostate cancer progression and metastasis.
CRISPR Medicine News reached out to Maria Rodrigo-Faus and Alvaro Gutierrez-Uzquiza for a comment on PRMT7, but they did not respond.
Link to Juan Martín Arriaga's original article in Oncogene:
Link to Martin Kristian Thomsen's original article in Nature Communications:
Link to Maria Rodrigo-Faus' original article in Cancer Letters:
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.