CRISPR-Select Validates Your Drug Target and Patient Group
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Many genetic variants can contribute to heritable cancers in, for example, the breast or ovary, but how most patient mutations contribute to disease is unknown. In his search for the mutations that cause the cancer, Claus Storgaard Sørensen, associate professor and group leader at the Biotech Research and Innovation Centre (BRIC) at the University of Copenhagen, and collaborators at Rigshospitalet, had sequenced blood cells from very young women with familial breast cancer.
» We found several mutations, but there was a huge lack of knowledge and technologies to determine whether the mutations had a harmful effect or not,« Sørensen recalls. He speculated a lot about it and shared his thoughts with his colleague Morten Frödin, who is also an associate professor and group leader at BRIC. With Sørensen's primary focus on the heritability of cancer and Frödin's interest in genomic engineering, they collectively devised a solution to the problem: CRISPR-Select.
CRISPR-Select is a fast and easy way to determine any genetic sequence variant's pathogenicity and drug responsiveness. In addition, the method is highly reliable because it controls for clonal variation, CRISPR off-target effects, false negatives, and other experimental confounders. CRISPR-Select was presented in December 2022 in a paper in Nature Genetics, and it is now the core, proprietary technology of the start-up company BioPhenyx, where the two scientists are co-founders.
All experiments are carried out in one pot
The basic principle of CRISPR-Select is to introduce an experimental genetic variant of a gene of interest in a cell culture and to compare its functional effect with a neutral control variant at the very same location. The experimental mutant could, for example, be changing the codon AAT (Asn) to AAA (Lys), while the same codon is neutrally changed to AAC (Asn) in the control (see Figure 1).
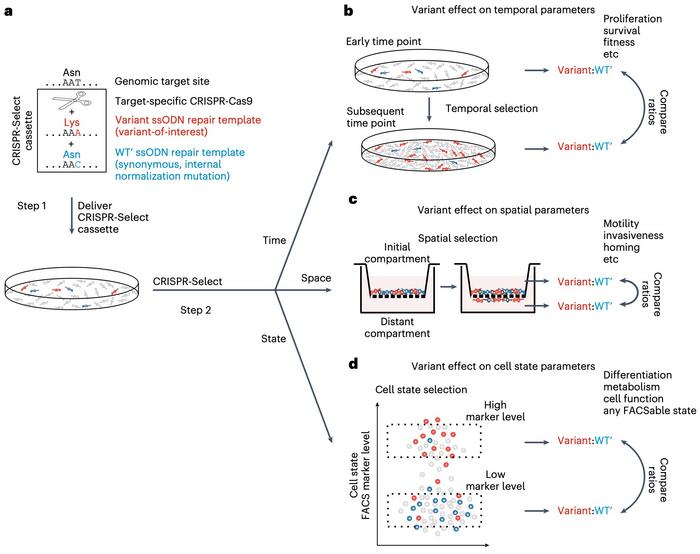
The two variants are introduced simultaneously in the same pool of cells by transfection with Cas9, a gRNA and two single-stranded oligodeoxynucleotides (ssODN) repair templates. The gRNA directs Cas9 to make a double-strand break (DSB) as close as possible to the base pair one wants to mutate.
“The mixed population of cells is then grown for ten days, and the same simple procedure is repeated - isolation of genomic DNA, one PCR reaction, and one NGS analysis. Then the new ratio of mutant and control cells is calculated, and the role of the mutant can thereby be conclusively determined”Morten Frödin
When the cells subsequently repair the DSB by homology-directed repair (HDR), the ssODNs will randomly direct the introduction of either the mutant or the control variant into some of the cells.
In the second step of CRISPR-Select, this pool of mixed cells is grown together under the same conditions. An aliquot of the cell population is taken after DNA repair is completed around day 2, and genomic DNA is isolated from the mixed pool of cells.
»Then we make a single PCR around the target site that amplifies all target site alleles present in the population of cells. Finally, the PCR products are sequenced by next-generation sequencing (NGS), so we obtain the absolute frequency of all the target site alleles,« explains Frödin.
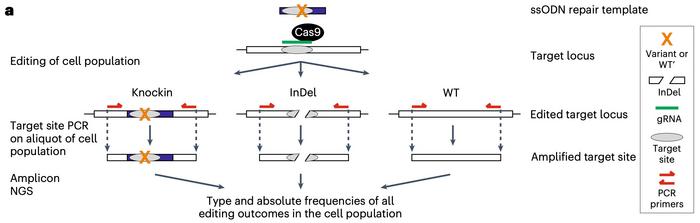
Oncogenic and tumour-suppressing variants are easily identified
In one experiment, Frödin elaborates, they obtained 2% mutant and 4% control cells. The rest of the cells were either unaffected or had other mutations, for example, indels introduced by non-homologous end joining (NHEJ) repair. The ratio of mutant and control cells is then calculated, and this is the only significant number to remember.
»The mixed population of cells is then grown for ten days, and the same simple procedure is repeated - isolation of genomic DNA, one PCR reaction, and one NGS analysis. Then the new ratio of mutant and control cells is calculated, and the role of the mutant can thereby be conclusively determined,« says Morten Frödin. In this particular experiment, mutant cells had become numerous, while control cells had remained at the initial level, leading to a much higher mutant:control ratio than at day 2. The firm conclusion that could be drawn from the experiment is that the mutation promotes cell proliferation or survival.
Sørensen, Frödin and co-workers tried CRISPR-Select in two experiments with known driver mutations in breast cancer to show proof-of-concept. In one experiment, the mutant ssODN repair template introduced a gain-of-function H1047R variant in the oncogene PIK3CA, while the control template introduced a silent variant at the same locus. In the other experiment, a loss-of-function T2722R variant and a silent control variant were introduced in the tumour suppressor gene BRCA2.

In both experiments, the mutant:control ratio at day 2 was close to 1 - indicating that the mutant and control templates had repaired the DSBs at equal frequencies - but these ratios changed dramatically over time, and they did so in opposite directions.
At day 12, cells with the mutant gain-of-function variant of the oncogene PIK3CA outnumbered cells with the control variant with a ratio of 13:1, demonstrating the aggressive carcinogenic effect of the mutation. On the other hand, cells with the loss-of-function T2722R variant of the tumour suppressor gene BRCA2 were only present at a ratio of 1:5 compared with the control variant on day 12. BRCA2 encodes a key factor for DNA repair essential for proliferating cells, and CRISPR-Select confirmed this.
CRISPR-Select suggests both the diagnosis and treatment
Mutations in BRCA2 and the related BRCA1 can, however, ultimately elicit cancer because BRCA-deficient cells cannot repair cancer-promoting mutations in genes such as PIK3CA. About 69,000 variants of BRCA1 and BRCA2 have been identified through diagnostic DNA sequencing in patients, but only 4,900 of these are known to be harmful, while 1,350 are classified as benign. Thus, the clinical significance of ~90% of all the variants is still unknown.
“In our experiments with PARP inhibitors, we can hardly find any surviving cells with the harmful variants - they disappear”Claus Storgaard Sørensen
However, the inventors of CRISPR-Select demonstrated that their method could distinguish between known benign and known pathogenic variants because the benign variants retain the same ratio of mutant to control from day 2 to day 12. In contrast, this ratio drops substantially over time for the pathogenic variants.
Moreover, CRISPR-Select can screen pathogenic BRCA variants for their response to a class of drugs called PARP inhibitors. Cancer patients with such variants often benefit significantly when treated with PARP inhibitors, while patients with non-responsive variants do not. When CRISPR-Select was used with or without the addition of the PARP inhibitor talazoparib to the growth medium, it was shown that while the drug strongly inhibited the proliferation of pathogenic BRCA variants, benign BRCA variants were unaffected.
»Cancer clinicians are looking for functional assays that distinguish benign and harmful variants and demonstrate which of the latter respond to drugs. In this respect, our assay is exceptionally effective. For example, in our experiments with PARP inhibitors, we can hardly find any surviving cells with the harmful BRCA variants - they disappear,« says Sørensen.
“In this way, we can normalise out all off-target effects, other unspecific CRISPR effects, and transfection toxicities. Thereby, any functional difference between cells with the mutant and control variants can be attributed to the mutation”Morten Frödin
Frödin adds that CRISPR-Select stands out from the competition because of the unique way the method generates and quantitates the control and mutant variants:
»Our control variant is functionally a wild-type which is generated in exactly the same way as the mutant variant, including using the same CRISPR reagent. In this way, we can normalise out all off-target effects, other unspecific CRISPR effects, and transfection toxicities. Thereby, any functional difference between cells with the mutant and control variants can be attributed to the mutation.«
These experiments show that CRISPR-Select can be used to optimise both the diagnosis and treatment of cancer. But the new method has even more potential.
All applications mentioned so far refer to what the authors classify as CRISPR-SelectTIME since the mutant and control variants are differentiated by their ability to proliferate or survive over time. However, two more versions of the method look at the variants' different abilities to perform on other parameters (see Figure 1C-D).
Clinicians are testing CRISPR-Select on patients
CRISPR-SelectSPACE is designed with cell motility as the differentiating parameter and can determine the cell's homing potential or ability to invade new compartments. In one experiment, control and mutant H1047R variants of the oncogene PIK3CA were introduced into cells placed on top of a transwell filter with epidermal growth factor (EGF) as a chemoattractant below. Mutant variants showed an enhanced ability to migrate into the lower chamber where the ratio of mutant:control cells had increased the next day.
“The clinicians have BRCA2 variants from patients but do not know if they are harmful or benign. Therefore, they will use CRISPR-Select to test if the variants are harmful and if they can be treated with PARP inhibitors”Claus Storgaard Sørensen
The method's third version, CRISPR-SelectSTATE, differentiates cells on, e.g., their cell cycle state, metabolism, or any state that can be tracked with fluorescence-activated cell sorting (FACS) markers. The potential of this version was demonstrated by generating control and mutant H1047R variants of PIK3CA. The researchers then marked the cells in the S-phase cell state, where cells are duplicating their genomes in preparation for cell division. The experiment showed that S-phase positive cells were enriched in PIK3CA mutant variants and thus demonstrated that the variant stimulated cell proliferation.
A similar FACS experiment revealed that cells with the PIK3CA mutant H1047R variant were enriched in the apoptosis-negative cell population. This demonstrates that the variant confers resistance to apoptosis.
Sørensen and Frödin work with clinical oncologists and geneticists at Rigshospitalet to validate CRISPR-Select on actual cancer patients. Sørensen explains:
»The clinicians have BRCA2 variants from patients but do not know if they are harmful or benign. Therefore, they will use CRISPR-Select to test if the variants are harmful and if they can be treated with PARP inhibitors.«
The clinical work has only started recently, and there are no results so far, but the clinicians are positive. In the meantime, Frödin and Sørensen continue to improve CRISPR-Select on behalf of BioPhenyx. One of their primary goals is to make it even more scalable, so they can eventually test all the 68,962 BRCA1 and BRCA2 variants and contribute to fighting cancer.
Link to the original article in Nature Genetics:
Multiparametric and accurate functional analysis of genetic sequence variants using CRISPR-Select
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







