CRISPR-StAR Enhances In Vivo Genetic Screening Precision
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
In vitro genetic screening using CRISPR-Cas9 has been instrumental in identifying essential genes across cell types, especially in cancer research. However, the predictive power of these findings for therapeutic applications is limited, as the simplistic conditions of two-dimensional (2D) cell cultures fail to replicate the complex microenvironment of tumours in vivo. Tumour development is influenced by diverse factors, including immune interactions, nutrient gradients, and extracellular matrix components, which are absent in vitro.
Furthermore, current in vivo approaches suffer from noise introduced by low cell engraftment rates, bottlenecks during tumour establishment, and clonal heterogeneity, reducing the robustness of genetic findings. To address these challenges, researchers in Austria sought to develop a screening method capable of maintaining high resolution and accuracy within complex in vivo environments.
Today in Nature Biotechnology, they describe a new method, CRISPR-StAR (Stochastic Activation by Recombination), that relies on generating internal controls within single-cell-derived clones by activating sgRNAs in a subset of cells while keeping others in a wild-type state (see Figure 1). This is achieved using a CreERT2 recombinase system to induce recombination at specific loxP and lox5171 sites on sgRNA constructs, creating mutually exclusive active and inactive populations within the same clone.
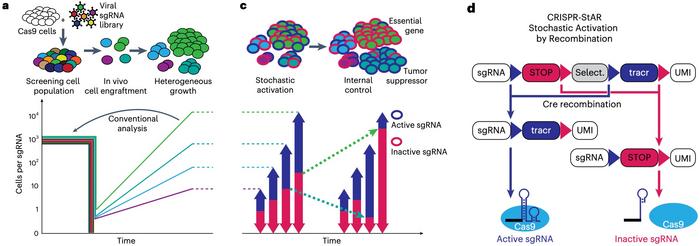
The design ensures that experimental and control cells share identical microenvironments and intrinsic conditions, eliminating variability from external factors. Unique molecular identifiers (UMIs) barcode each cell, allowing precise tracking of clonal behaviour. This intrinsic control approach drastically reduces experimental noise, enabling reliable hit calling even in sparse and heterogeneous data conditions, such as those encountered during tumour engraftment.
When tested in therapy-resistant mouse melanoma models, CRISPR-StAR consistently outperformed conventional CRISPR screens, especially under conditions of low cellular representation. Genome-wide screening revealed specific in vivo genetic dependencies, including mitochondrial genes critical for oxidative phosphorylation, which were not identified in vitro. These results highlight the method’s ability to uncover genes uniquely relevant to the tumour’s physiological context.
Benchmarks showed improved reproducibility, enhanced statistical power, and reduced false positives, even at minimal cell coverage. Subsequent validation experiments confirmed the essential roles of several identified genes, including some with potential therapeutic relevance. By faithfully modelling tumour biology, CRISPR-StAR provides a transformative tool for high-resolution genetic screening in vivo.
The study, conducted by Ulrich Elling’s team at the Institute of Molecular Biotechnology, Austria, and collaborators from Vienna and Toronto, was published today in Nature Biotechnology.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.