Editing Stem Cells In Vivo: A Major Stride in Gene Therapy for Blood Disorders
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Significant progress in gene editing over the last decade has offered hope for the treatment of many genetic disorders, but there remain key obstacles in the treatment of most inherited conditions.
For blood disorders, such as sickle cell disease (SCD), the target cells for gene editing are haematopoietic stem cells (HSCs), which reside in the bone marrow. Their location has made them difficult to target; until now, they had to be removed from the body and edited ex vivo before being transplanted back into the patient.
Despite several ongoing clinical trials for ex vivo gene-edited therapies to treat SCD and other diseases, there are two key disadvantages to this approach. Firstly, ex vivo therapies are expensive, costing between two and three million dollars per treatment. Secondly, to allow the edited cells to engraft, the diseased cells must first be eliminated – usually with gruelling genotoxic chemotherapy regimens.
In a study published last month in Science, researchers from the University of Pennsylvania (UPenn) have circumvented these obstacles, by using mRNA technology to edit HSCs in vivo with high specificity. Their approach offers critical solutions for genetically-modified autologous HSC therapy, as well as allogeneic donor HSC transplant.
»We are hopeful that in vivo editing will make gene therapy a more affordable and accessible option for patients moving forward, and it’s proof-of-concept that this can be done for other cells as well,« said Hamideh Parhiz, a research assistant professor of medicine at UPenn and senior author of the study.
The need for in vivo editing and how to target HSCs
So how did Parhiz and her colleagues manage to target HSCs in vivo? Firstly, they looked for highly specific surface markers that were expressed on the target cells. By generating antibodies targeted against these markers and covering lipid nanoparticle (LNP) delivery vehicles with them, they could test how well each antibody was able to target HSCs in whole bone marrow in vitro.
»When we compared different antibodies against various HSC surface markers, we identified CD117 as a good target, we saw that the editing efficiency that we got with CD117 was the highest. And that's why we went with it – it’s considered a universal HSC marker,« Parhiz explained.
Then, they packaged the mRNA cargo into the LNP: cyclic AMP response element (Cre) recombinase and a CRISPR-Cas9 adenine base-editor fusion gene.
They tested the system in vitro first, by isolating HSCs from mice, editing them ex vivo, and then injecting the edited cells back into mice. By performing cell lineage tracking, they were able to show that the transplanted cells were true HSCs, meaning that they were able to differentiate into multiple cell lineages, even after secondary and tertiary transplants.
Sickle cell disease: restoring haemoglobin expression with base editing
Once the team had established the targeting method and the long-term multipotency of the target cells in vivo, they wanted to show proof-of-concept in a relevant disease model. For Parhiz, choosing which disease to target was a no-brainer.
»When you’re talking about manipulating stem cells, one of the first questions is if you can do anything for diseases such as sickle cell disease, right? Co-senior author Stefano Rivella’s team actually had access to cells coming from sickle cell patients, and so we showed that we could edit those cells in vitro,« she said.
With HSC specimens from four different SCD donors, the team used the CD117-LNP system to deliver an mRNA cargo consisting of a Cas9 adenine base-editor (ABE) fusion along with a sgRNA targeting the disease-causing mutation in the beta-globin (HBB) gene to the cells.
A simple A-to-G substitution with the ABE system converted the pathogenic sickle mutation (HBBS) to the naturally occurring, non-pathogenic Makassar variant (HBBG). This strategy was able to almost eliminate the sickled cells, and did not affect the viability or proliferation of erythroid progenitor cells.
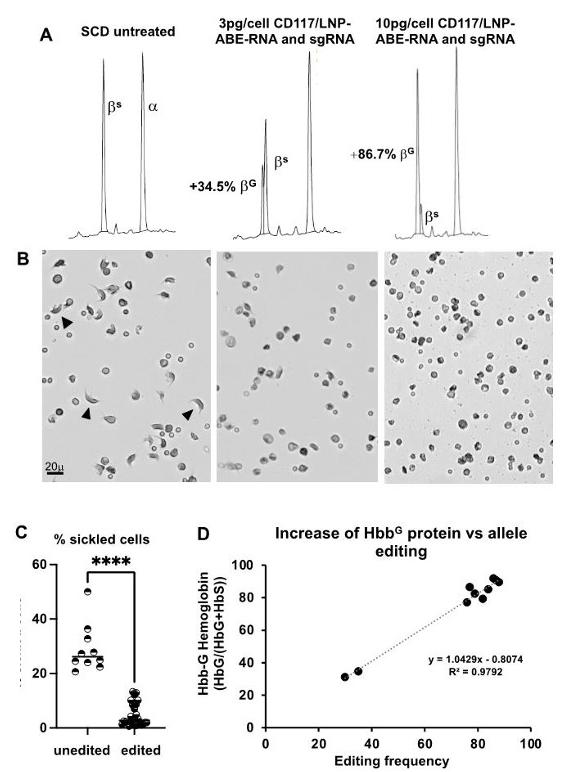
Base editing of the E6V sickle cell mutation with human CD117 targeted LNP. (A) Representative RP-HPLC chromatograms of in vitro-differentiated SCD erythroid progenitor lysates untreated (L) and after treatment with anti-human CD117(hCD117)/LNP-NRCH Cas9 ABE-8e mRNA and hCD117/LNP gRNA (middle and R). Base editing yields non-pathogenic HBBG (βG), which elutes before pathogenic HBBS (βS) and the α-globin protein (α). (B) Representative images of sickling of in vitro-differentiated erythroid progenitors under hypoxic conditions at the treatments in (A). Arrowheads indicate sickled morphology. (C) Percentage of sickled cells from unedited and edited sickling assays. (D) Correlation of %βG by RP-HPLC (protein) to base edited allele frequency (DNA).
An unrelated attempt to circumvent the need for ex vivo cell therapy for SCD was made by researchers earlier this year. An article published in Blood, which includes Professor David Liu of the Broad Institute as co-author, demonstrated proof-of-concept of in vivo HSC editing in a mouse model of SCD.
However, the strategy developed in that earlier study varies considerably from Parhiz’s: those authors used a helper-dependent adenoviral vector (HDAd) to deliver a prime-editing (PE) system to correct the sickle mutation rather than convert it to the Makassar variant. They also targeted CD46, a receptor expressed on primitive HSCs, rather than CD117.
Furthermore, unlike the present study, the HDAd system was unable to target the HSCs in vivo within the bone marrow and required subcutaneous injections of granulocyte-colony stimulating factor (G-CSF)/plerixafor (AMD3100) to mobilise the HSCs into the peripheral blood.
Parhiz and her colleagues’ approach to human SCD was only tested in vitro, however combined with the proof-of-concept of in vivo HSC editing, it could offer a promising alternative to current expensive cell therapy options for diseases that require genotoxic conditioning.
In vivo editing enables healthy donor HSC transplant
In addition to the successful editing of sickled HSCs, the paper also addresses treatment obstacles for another disease. Patients with severe combined immunodeficiency (SCID) can be treated with HSC transplant (HSCT), however, they are often unable to tolerate the extreme toxicity of the chemotherapy required to create space for the donor cells to engraft. These genotoxic treatments also put SCID patients at increased risk of developing secondary malignancies.
»[These therapies] have significant side effects - most of them can make the patient infertile. In this specific case, we are talking about paediatric patients, so this is very significant,« Parhiz said.
Parhiz and her team reasoned that if they could therapeutically edit HSCs in vivo with high specificity, they may also be able to ablate the HSCs in a specific manner, creating a niche for healthy donor cells to engraft while eliminating the need for genotoxic chemotherapy in SCID patients.
HSCs depend on the expression of anti-apoptotic gene MCL-1, so to ablate them, the team tried using their CD117-LNP system to deliver an mRNA cargo of a pro-apoptotic gene known as PUMA (p53 up-regulated modulator of apoptosis) to mice in vivo. However, they soon discovered that this would be one of the most challenging aspects of the study; the PUMA cargo was highly toxic and caused significant liver damage in the mice.
»The way we solved this problem was that we added specific sequences, called micro-RNA target sites, to the 3-prime untranslated region of the mRNA cargo. These sequences suppress the mRNA expression in the liver,« Parhiz explained. »That’s when we started seeing the effects – we could kill the HSCs and we didn’t have any toxicity.«
Once they had solved the toxicity issues, the team were able to demonstrate that their method enabled the successful transplant of bone marrow, with engraftment rates that are high enough for SCID to be cured – an enormous success.
Moving forward: assessing safety and lowering the cost of treatment
The findings presented in the current study are promising, but at this early stage, Parhiz says it’s hard to give an accurate estimate of the cost of any resulting therapies. However, the main elements needed have already been successfully scaled-up with good manufacturing practise requirements at relatively low costs – for example, in the context of COVID-19 mRNA/LNP vaccines.
»We think that it will be cheaper. We are also hopeful that the financial burden on the health system would also be less because these injections can be done by any nurse or physician. This doesn't require hospitalisation, or the type of infrastructure that's needed for cell therapy. So I think, yes, hopefully, it will be cheaper,« Parhiz remarked.
Professor Donald Kohn of the University of California Los Angeles, who was not involved in the study, said it’s a major step in the right direction. »Basically, in vivo gene delivery is the Holy Grail for the field of HSC and blood diseases,« said Kohn, who specialises in the treatment of these same disorders. »This paper using lipid nanoparticles presents promising preliminary data to show efficacy in a mouse model.«
There are many more potential considerations to be addressed before the therapy can progress toward clinical trials. »Issues will be the efficacy for more difficult cargoes, the scale-up to human subject size, and issues of specificity of the delivery,« Kohn added.
Because Parhiz and her colleagues originally aimed to reduce or eliminate the treatment risks for patients, they are also determined to make these therapeutic avenues as safe as possible.
»Because this has potential in paediatric applications, we need to make sure that we are dealing with a system that has a really good safety profile. So, we need to look carefully at the toxicity profile in humanised mice models and non-human primates, or other larger animal models. After that, we are hopeful [that we can progress to the clinic] and we will move forward.« Parhiz concluded.
Link to the original article in Science:
In vivo hematopoietic stem cell modification by mRNA delivery
Rebecca Roberts is a molecular biologist, science writer/communicator, and scientific marketing professional based in Queensland, Australia.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsIn vivoSevere Combined Immunodeficiency, SCIDSickle Cell Disease, SCDBase editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.