Engineered Probiotic Delivers CRISPR-Cas9 Kill Module To Antibiotic-Resistant Pathogens in Mouse Gut
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
The treatment of bacterial infections is contingent on the efficacy of current antibiotic therapy, but this is seriously threatened by the rapidly increasing problem of antibiotic resistance.
The U.S. Centers for Disease Control and Prevention estimate that more than 2.8 million antibiotic-resistant infections occur yearly in the U.S., while the European Centre for Disease Prevention and Control reports more than 670,000 resistant infections annually in the EU, with estimated health-care costs of around 1.1 billion euros. In short, new ways to fight antibiotic resistance are urgently needed.
Bacterial conjugation to deliver CRISPR systems has been explored as an approach to target antibiotic-resistant bacteria. This relies on the ability of certain bacteria to conjugate with a donor cell to which it can transfer DNA. Despite modest early successes with this approach, poor transfer rates have created a bottleneck to its widespread success.
In a recent study published in Molecular Systems Biology, researchers from Dr Sébastien Rodrigue’s research group at the Université de Sherbrooke, Québec, Canada, have removed that bottleneck. They describe a novel conjugative probiotic strain of E. coli that can propagate a CRISPR-Cas9 system with high transfer rates within the gut microbiota of mice. They demonstrated that their new system could clear an antibiotic-resistant gastrointestinal infection in mice, providing hopes for new ways to treat antibiotic-resistant infections.
Commenting on this importance of this study, Dr Rodrigue says: »This work shows that conjugation can reach higher rates, and that it can be used for microbiome editing by making the delivery sufficiently good to reach most of the cells that you want to eliminate and clear an infection«.
Improving conjugation rates is key
Bacterial conjugation to deliver CRISPR-Cas constructs in lab cultures was first explored in an early study in 2019 by researchers at The University of Western Ontario in Canada, followed by another study later that year by researchers in Finland and the UK. Other groups have explored conjugative interference or conjugative CRISPR-Cas9 plasmids for use in mouse models, but poor transfer rates have limited the success of these approaches.
Rodrigue’s team started out with microbial community experiments to assess the efficacy of using a conjugative probiotic to deliver a CRISPR-Cas9 system in vitro. Building upon an earlier study, where they screened conjugative plasmid families in enterobacteria and identified plasmid TP114 as an efficient DNA transfer machinery in the mouse gut microbiota, they first integrated a CRISPR-Cas9 module in the TP114 conjugative plasmid. They then introduced the resulting construct into a donor conjugative probiotic (COP) E. coli strain.
The team then co-cultured the COP in vitro on agar plates, with a target and a control E. coli strain, each of which was engineered to carry a distinct antibiotic-resistance gene in its genome. These antibiotic-resistance genes were subsequently used as genomic CRISPR targets (Figure 1). Since each strain carries a specific antibiotic-resistance gene, sampling and culturing the bacteria on selective agar allowed the researchers to easily identify each strain and quantify the viability of the target strain after conjugative probiotic treatment.
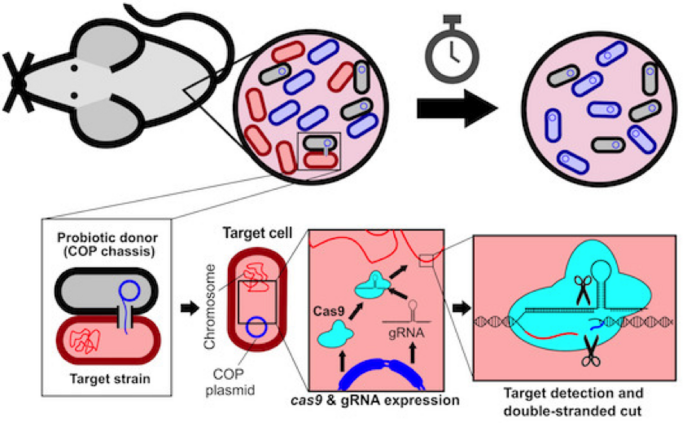
By counting the number of colony-forming units (CFUs) present on agar cultures 24 hr after conjugative probiotic treatment, the team found that the construct was efficiently disseminated throughout the population, specifically by observing killing of the target strain while leaving control strain levels intact.
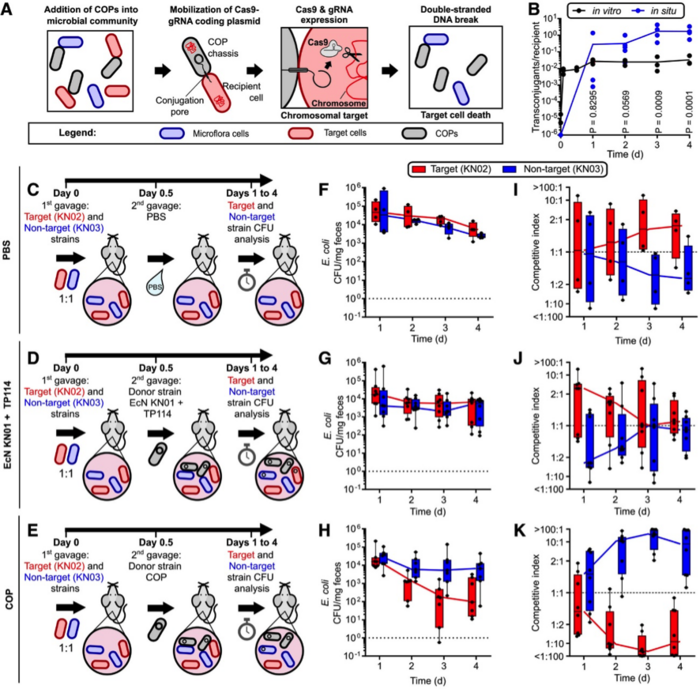
Next, the activity of the COP strain was assessed in the mouse gut microbiota. Mice were colonised with the target and control strains (introduced by gavage), followed by administration of the COP and measurement of the competitive index (ratio of absolute CFU) between both strains. The system proved to be highly efficient: COP treatment resulted in elimination of 98.6% of the target inoculum after 36 hr, and elimination of 96.4% when the COP was administered as a prophylactic treatment 12 hr before target and control strain introduction (Figure 2).
Improving pathogen killing via adaptive laboratory evolution
Excited by the positive outcome, the group wanted to go even further, as Dr Rodrigue outlines: »We wondered if we could make this system work even better. Killing almost 99% of the bacteria is good, but if you have a million bacteria per milligram of faeces, a lot of bacteria are still alive, and we wanted to push the limits of the system.«
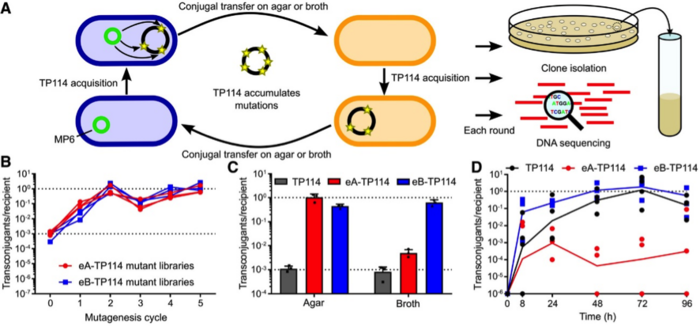
The team designed an adaptive laboratory experiment to generate a highly-improved TP114 plasmid, as Dr Neil explains: »High mutation rates were promoted within the donor strain to create a library of TP114 mutants, and these mutants then competed with one another in several conjugation cycles. In each cycle we induced additional mutations and selected through conjugation the ones that transferred faster. It is natural selection coupled with accelerated evolution. We then introduced the CRISPR-Cas9 module into the improved TP114 plasmid.«
The improved conjugation system did not disappoint – the target strain population was reduced by 95% within only 12hr after COP administration, and this rose to 99.5% after 2 days, and 99.99% after 4 days (Figure 3).
David Edgell, Professor in the Department of Biochemistry at The University of Western Ontario in Canada, who has explored gene-editing tools and synthetic biology to control microbial populations, but who was not involved in the current work, sees great potential in using plasmid conjugation for microbiome editing:
»The use of laboratory evolution to select plasmids with higher rates of conjugation that are relevant for in vivo applications highlights the potential of conjugative delivery of CRISPR nucleases to eliminate specific bacteria within complex communities. It will be interesting to monitor conjugation rates and plasmid stability in complex microbiomes long term, as optimising these factors will be crucial for many in vivo applications.«
Co-opting the bacterial conjugation system
Bacterial conjugation is often seen as a sub-optimal delivery system due to traditional low transfer rates. This is in stark contrast to the high infectivity of bacteriophages, which are widely explored as DNA delivery systems for novel antimicrobials today.
According to Dr Rodrigue, however, a conjugation-based system may represent a superior alternative to traditional bacteriophage delivery: »One advantage of conjugation is that the bacteria can continue to deliver the CRISPR system over again. When you have a phage, it is pretty sensitive to different environmental conditions, and it will only inject DNA in one cell, whereas the bacteria will continuously deliver the DNA.« In other words, the COP system developed in Dr Rodrigue’s lab allows for greater efficiency, as cells that receive the plasmid via conjugation can in turn disseminate the CRISPR module themselves.
In the future, the group intends to study the host range of their system to ascertain whether additional CRISPR modules should be constructed, as Dr Rodrigue explains: »TP114 disseminates pretty well to E. coli and other enterobacteria, but it is not clear if it will disseminate well to more phylogenetically distant targets.« The team concedes that if TP114 is unable to conjugate with other bacteria, they may have to rebuild the system for other bacterial pathogens and make sure that the different conjugation machineries provide sufficient host range.
Dr Rodrigue points out another important consideration for studies that use bacteria as the delivery vehicle: » In studies such as this, it is necessary to protect the donor strain from its own CRISPR cargo. E. coli strains have differences in their genome. To design a spacer that targets all E. coli strains in a highly conserved [genome] region, you need to recode that region of the genome in your donor strain to prevent it from killing itself.«
Complete pathogen clearance in a simulated gut infection scenario
Before any technology can progress to human clinical trials, animal model testing is a paramount step. Differences in microbiome composition and complexity between mouse infection models and human infections can hinder knowledge translation.
To better simulate a real infection scenario, the team administered their conjugative probiotic in mice colonised with the gut pathogen Citrobacter rodentium, without previous antibiotic treatment of the microbiota to deplete enterobacterial populations. This is highly relevant since the C. rodentium mouse infection model is often used to simulate human gut infection by E. coli strains.
Remarkably, the COP was able to clear 99.98% of the C. rodentium inoculum within just 4 days of treatment, providing two important pieces of data: the conjugative probiotic carrying a CRISPPR-Cas9 system can target other enterobacteria, and it can be used to clear an infection in a context that more closely resembles a human clinical setting.
This finding is even more incredible considering that the control C. rodentium infection could not be fully cleared within 4 days of standard antibiotic (streptomycin) treatment, an observation that hinted to the emergence of resistant cells, while the COP strain carrying the CRISPR system achieved complete clearance of Citrobacter rodentium without the detection of resistance. This increases the promise of the CRISPR-Cas9 system to target an antibiotic-resistant infection that would not be phased by conventional antibiotic therapy.
Swine model is a major stepping-stone to human trials
Highly encouraged by their results so far, Rodrigue’s team has already initiated similar experiments in swine. Dr Rodrigue explains the rationale behind this: »The digestive system of pigs is much more similar to humans compared to the mouse. If the system works with swine, then there is a very high chance that it will work in humans.« Swine trials also provide important clues to improve agriculture applications, since antibiotics are heavily used during piglet development to prevent infection. The success of the conjugative probiotic approach could also help to decrease antibiotic load in animal farming.
Although swine studies will help to answer many important questions about the feasibility of COP-based therapy in humans, there are still several unknowns that will need to be addressed, as Dr Rodrigue highlights: »Nutrition is another factor with an impact in microbiome composition that may be difficult to control in future human clinical trials, but this is not the only confounding factor. Even if food was completely controlled, there are other components like genetics or life history that will have an important role in microbiome composition. Some people might react better to this type of treatment depending on the composition and robustness of their microbiome. That is something we don’t know much about.«
Safety issues must be addressed before clinical use
The group is mindful of safety issues, especially considering the use of genetically-modified organisms. The use of kill switches or auxotrophies has been studied as biocontainment methods for modified organisms. Following up on this aspect, Dr Rodrigue’s team wants to constrain the dissemination of the plasmid or the engineered probiotic, since there is a higher chance that resistance mechanisms may evolve if the system spills into the environment. Dr Rodrigue outlines the plan to mitigate this risk: »The strain to use in a future in clinical application would have a kill switch or an auxotrophy that would make it die after it is out in the environment.« So far, the team has performed preliminary experiments with the dapA gene (lysine biosynthesis), which is commonly used as an auxotrophic marker.
Recognising that even if the engineered probiotic dies, there is still a chance that the plasmid could survive and move to other bacteria, Dr Rodrigue presents two possible solutions: »One would be to just deliver the CRISPR reagents, but without the conjugation machinery, so there would be only one donor bacterium and everything else would be a recipient. The other approach would be to use two sets of guide RNAs: the first guide(s) would cleave the target genome, while the second guide would cleave the [CRISPR] plasmid and make sure it is destroyed.«
Fresh hopes for antibiotic resistance and chronic disease
A prime driver for the development of CRISPR-based approaches is to fight antibiotic resistance. Dr Rodrigue discusses one of the most likely approaches – using antibiotics and CRISPR antimicrobials in conjunction – »There is a strong rationale for using both: use the antibiotic to kill what is susceptible and if there is a resistant population of cells, then you will have the engineered probiotic to sensitise them.«
Since they developed a system capable of clearing an infection by direct killing of the pathogen, Dr Neil believes it could someday be used as a stand-alone therapy:
»Any antibiotic-resistant infection could be tackled with the COP technology, it could be better to just kill the bacteria«. He also reflects on other applications ahead: »The treatment can be prophylactic. If you await surgery, you could take the bacteria [COP] to lower the antibiotic resistance profile of any bacteria you may be harbouring and to prevent any chance of developing an antibiotic-resistant infection.« On that note, the team is also interested in exploring other body niches, including the urinary tract or the skin.
Dr Rodrigue also sees the technology as a way to rebalance the microbiome in patients with chronic disease: »A nice application of our system would be to try to re-establish a healthy microbiome for diseases where there is a microbiome component. For instance, we know that people that suffer from Crohn's disease often have very high levels of enterobacteria and inflammation. It's not clear if inflammation promotes the rise of enterobacteria or the opposite. There could be applications in the clinic for these conditions, or more fundamentally to help us understand the sequence of events.«
Link to the original article in Molecular Systems Biology:
High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing
Pedro Pais is a molecular biologist and microbiologist based in Portugal.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsDeliveryNon-viralInfectious diseaseMicrobiomeCRISPR-CasCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







