Highlight: CRISPR-Edited RUNX1 Variants Enhance Osteogenesis
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Bone mineral density (BMD) is a complex trait influenced by genetic, behavioural, and environmental factors, with heritability estimates reaching up to 80%. Recent genome-wide association studies (GWAS) have identified over 500 loci associated with BMD, including intronic and intergenic variants near the RUNX1 gene, which is known for its role in skeletal development and repair.
However, how these non-coding variants regulate gene expression and influence osteogenesis remained unclear. The study aimed to uncover the functional roles of these variants in regulating bone formation and repair and pave the way for future therapies targeting bone diseases such as osteoporosis.
To dissect the regulatory roles of BMD-associated variants at the RUNX1 locus, researchers employed CRISPR-Cas9 genome editing to create human induced pluripotent stem cell (iPSC) models. Two deletion lines, Δ1 and Δ2, were engineered to excise regions containing these variants. These iPSCs were subsequently differentiated into mesenchymal stromal cells (iMSCs) and osteoblasts.
The effects of the deletions were evaluated using in vitro assays for osteogenic differentiation and in vivo mouse models for bone regeneration. Circularised chromosome conformation capture sequencing (4C-Seq) was performed to map chromatin interactions, revealing how the deleted regions interacted with RUNX1 and other genes, such as SETD4, involved in chromatin modification.
The experimental results demonstrated that the deletion mutant lines exhibited enhanced osteogenic potential compared to wild-type controls. In vitro, iMSCs derived from Δ1 and Δ2 lines showed increased mineralisation and upregulated expression of osteogenic markers like RUNX2 and SP7. 4C-Seq analyses revealed altered chromatin interactions, with the deletion mutants disrupting a regulatory hub, leading to increased expression of RUNX1 and SETD4.
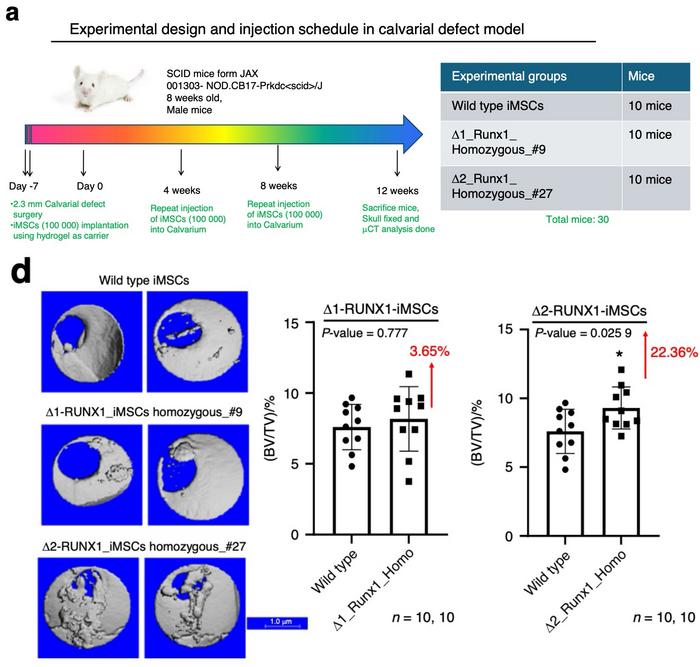
In vivo studies utilising a mouse calvarial defect model further validated the enhanced osteogenic potential of the CRISPR-edited RUNX1 deletion mutants (see Figure 1). Human mesenchymal stromal cells (iMSCs) derived from wild-type and deletion mutant (Δ1 and Δ2) lines were implanted into sub-critical-sized cranial defects in SCID mice. The Δ2 deletion line, in particular, demonstrated significantly increased bone formation compared to both wild-type and Δ1 mutants. This was evidenced by greater mineralisation and improved repair of cranial defects, as shown by micro-computed tomography (micro-CT) and histological analyses.
These results highlight the Δ2 deletion’s ability to stimulate osteogenic activity in vivo, aligning with its enhanced performance in vitro. The findings not only confirm the functional importance of the RUNX1 regulatory loci in skeletal biology but also suggest the potential utility of these gene-editing strategies in developing treatments for bone regeneration and repair.
The study was led by Nazir M. Khan and Hicham Drissi from Emory University School of Medicine, and it was published last Friday in Bone Research.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.