Hit-and-Run Epigenome Editing Provides Efficient and Durable Epigenetic Silencing In Vivo, New Study Shows
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The use of programmable editors equipped with transcriptional repressor domains has been proposed for the treatment of genetic diseases through permanent epigenetic silencing of specific genomic sites — this approach is referred to as epigenome editing. However, limited information is available on the translation of epigenome editing approaches from in vitro to in vivo settings, which is a key step in the development of new therapies. This has limited the development of epigenetic silencing as a potential treatment.
In a recent experimental validation study, researchers at the San Raffaele - Telethon Institute for Gene Therapy (SR-Tiget), Università del Piemonte Orientale, and Vita-Salute San Raffaele University used hit-and-run epigenome editing to target mouse Pcsk9.
Pcsk9 encodes a serine protease that is predominantly expressed in the liver and regulates cholesterol homeostasis by promoting degradation of the low-density lipoprotein receptor (LDL-R). In turn, LDL-R degradation results in higher plasma levels of LDL-C — also known as ‘bad’ cholesterol. Gain of function mutations in this gene can lead to hypercholesterolemia and atherosclerosis, making it an attractive target for therapeutic interventions.
»Our lab is keen on developing new tools to regulate gene expression without affecting genetic sequences, and in this study, we focused on silencing Pcsk9,« said Martino Alfredo Cappelluti, PhD, the first author of the study. »Although small molecules can effectively control hypercholesterolemia, they require life-long daily administrations and are associated with some toxicities. More advanced therapeutic approaches, such as monoclonal antibodies and RNA interference, require repeated administration.«
The study showed that one-time delivery of programmable editors in vivo using lipid nanoparticles (LNPs) resulted in durable epigenetic silencing of Pcsk9 in mice, even after forced liver regeneration. The researchers further improved the design of the engineered transcriptional repressor, which provided efficacy similar to that obtained with conventional gene editing.
This proof-of-concept study provides experimental evidence that transcriptional repressors may provide long-lasting epigenetic silencing of target genes without safety concerns associated with the introduction of DNA breaks into the genome, laying the groundwork for further development of epigenetic silencing-based therapeutics.
The study was published in Nature.
Zinc-finger proteins produce robust epigenetic silencing of mouse Pcsk9
Researchers conducted in vitro screening of different DNA-binding enzymes, including endonuclease-dead Cas9 (dCas9), zinc-finger protein (ZFP)-based engineered transcriptional repressors (ETRs), and transcription activator-like effectors (TALEs)-based ETRs. This screen aimed to evaluate the efficiency of different platforms in silencing mouse Pcsk9. The team used nucleofection to deliver mRNAs encoding the various editors into a mouse hepatocyte cell line that expresses a fluorescently tagged Pcsk9. As a control, cells were nucleofected with Cas9 mRNA and a gRNA targeting the first exon of mouse Pcsk9.
Evaluation of the efficiency of gene silencing with different ETR designs involved analysis of the percentage of Pcsk9-negative cells using flow cytometry. The researchers found that ZFP was the editor design that most efficiently silenced mouse Pcsk9 in hepatocytes (Figure 1). In a dose-response analysis, EC50 (half-maximum effective concentration) was 0.08 for ZFP-ETRs, 0.21 for TALE-ETRs, and 0.38 for dCas9-ETRs, highlighting the superior efficiency of ZFP-ETRs.
Although it remains unknown why ZFN-ETRs performed better than other editor designs, Dr Cappelluti speculated that their superior efficiency may be related to the fact that it is a compact system in terms of size and, thus, one can deliver more RNA of this species. »For CRISPR-based systems, you need to deliver multiple RNAs that express the editors and the guide RNA,« he explained.
The team also performed RNA sequencing analysis to assess the specificity of the ZFP-ETRs. In addition to a ~30-fold reduction in Pcsk9 levels after treatment with ZFP-ETRs, eight other genes were also deregulated (four upregulated and four downregulated), albeit to a lesser degree.
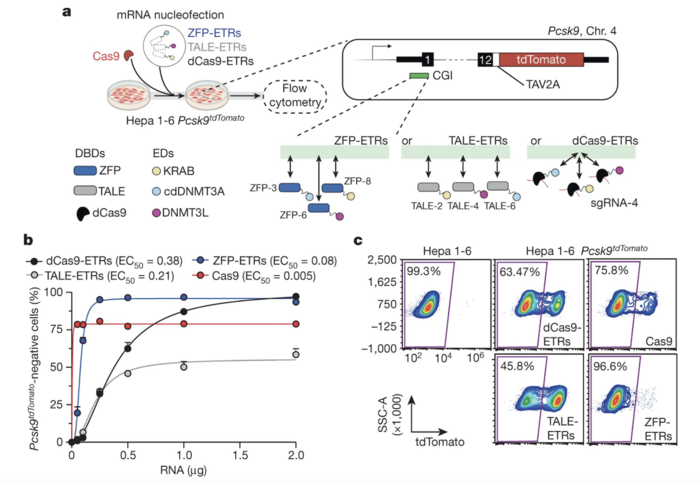
Figure 1. ZFP-based ETRs are the most effective platform for epigenetic silencing of mouse Pcsk9. (a) Schematic overview of in vitro screening comparing the efficiency of different ETR designs in mouse hepatocytes. (b) Percentage of Pcsk9-negative cells at day 13 after delivery of increasing doses of mRNAs encoding dCas9-, TALE-, and ZFP-based ETRs. (c) Representative flow cytometry plots 29 days after RNA nucleofection. Cappelluti et al. Nature (2024). https://doi.org/10.1038/s41586-024-07087-8
Single delivery of ZFP-based ETRs epigenetically silences Pcsk9 in the mouse liver
Researchers used LNPs to deliver the ZFP-based editing machinery to the liver of mice. Compared with PBS injection (control), transient delivery of ZFP-based ETRs via a single intravenous injection of LNPs loaded with editor mRNAs reduced circulating Pcsk9 levels by nearly 40% (Figure 2). Moreover, LDL-C levels decreased by 35% 30 days after injection of ETR-loaded LNPs. The levels of circulating Pcsk9 remained low for almost one year, suggesting that a single administration of ZFP-ETRs may provide efficient and sustained epigenetic silencing of mouse Pcsk9in vivo. Dr Cappelluti noted that increases in liver enzymes, indicative of treatment-related hepatotoxicity, were transient and likely caused by the LNP formulation, not the ZFN-ETRs.
To further validate the ability of this approach to promote long-term epigenetic silencing, the team performed partial hepatectomy three months after LNP injection to induce liver regeneration. The researchers found that decreases in circulating Pcsk9 levels and the levels of CpG methylation of the Pcsk9 promoter were similar before and after hepatectomy (Figure 2). This finding suggests that epigenetic modifications can persist through cell replication, confirming the heritability of epigenetic Pcsk9 silencing and the potential of this approach for long-term therapeutic effects without the need for continuous treatment.
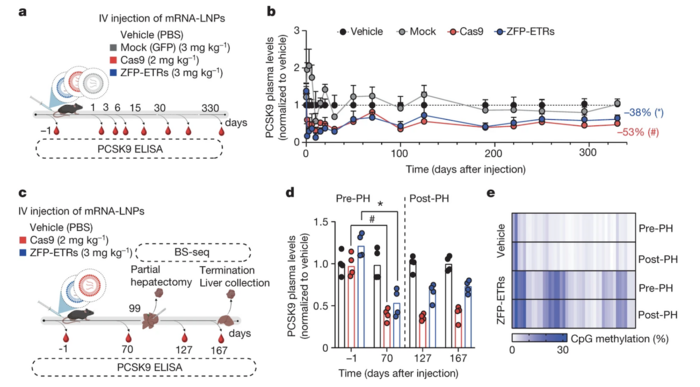
Figure 2. Epigenetic silencing of Pcsk9 in mouse liver after a single administration of ZFP-ETR–loaded LNPs. (a) Schematic overview of the experimental procedure used to assess the efficacy of Pcsk9 silencing in vivo. (b) Circulating Pcsk9 levels at different time points after LNP injection. (c) Schematic overview of the experimental procedure used to assess the durability of Pcsk9 silencing in vivo after partial hepatectomy. (d) Pcsk9 levels before and after partial hepatectomy. (e) Heatmap showing the average CpG methylation in Pcsk9 in ZFP-ETR–treated and control mice before and after partial hepatectomy. Cappelluti et al. Nature (2024). https://doi.org/10.1038/s41586-024-07087-8
EvoETR: An all-in-one design for epigenetic silencing of mouse Pcsk9
To further improve the efficiency of in vivo silencing of mouse Pcsk9 using ZFP-based ETRs, the researchers modified the editor design to produce an all-in-one construct — they termed this ‘evolved engineered transcriptional repressor’ or EvoETR. Combining multiple domains into a single RNA molecule offers several advantages, including reduced complexity, higher epigenetic silencing efficiency and specificity.
EvoETR-8, one of the all-in-one configurations tested, demonstrated substantial epigenetic silencing efficiency in vitro, producing strong and durable Pcsk9 silencing comparable to the triple-ETR combination (~85% Pcsk9-negative cells for both). RNA sequencing and genome-wide CpG methylation analyses showed that EvoETR-8 was highly specific, with comparable genome-wide methylation patterns between EvoETR-8- and mock-treated cells.
Moreover, EvoETR-8 showed high efficiency in epigenetically silencing mouse Pcsk9in vivo, reducing the circulating levels of Pcsk9 by 75% (Figure 3). The efficiency of EvoETR in reducing Pcsk9 levels in vivo was comparable to that of conventional gene editing, which led to a 70% reduction in circulating Pcsk9 levels. These two approaches also led to similar reductions in LDL-C levels (29% with EvoETR-8 and 34% with gene editing).
»The advantage of this approach over gene editing is that no DNA breaks are introduced, which is a key safety concern that limits the use of CRISPR-Cas9 in humans for therapeutic purposes,« noted Dr Cappelluti.
The study also showed that the epigenetic silencing platform is portable to other particle formulations, confirming its robustness and versatility for different delivery systems.
Commenting on the implications of these findings, Dr Cappelluti said, »EvoETR represents a major step forward in the design of safer and more effective gene editing tools.«
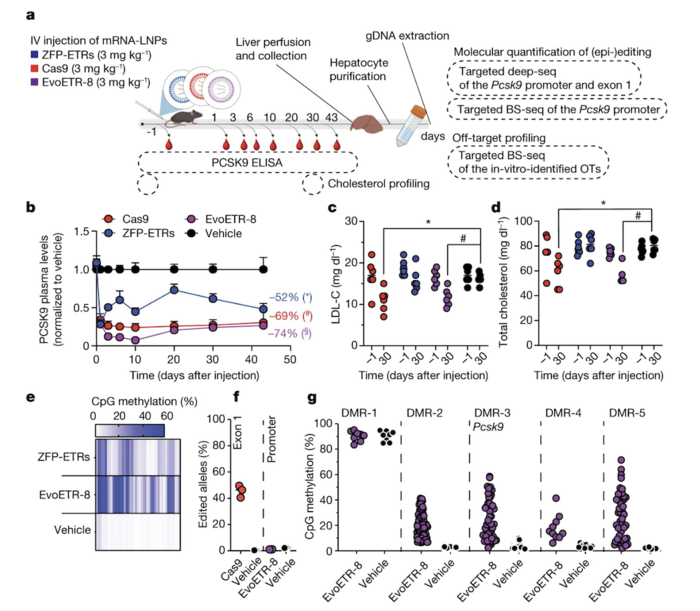
Figure 3. Epigenetic silencing of Pcsk9 after a single administration of LNPs loaded with EvoETR-8. (a) Schematic overview of the experimental procedure used to assess the efficacy of EvoETR-8 in vivo. (b) Circulating Pcsk9 levels at different time points after LNP injection. (c, d) Levels of LDL-C and total cholesterol 30 days after LNP injection. (e) Heatmap showing the average CpG methylation in Pcsk9 in treated and control mice. (f) Percentage of edited alleles. (g) Percentage of in vivo methylation. Cappelluti et al. Nature (2024). https://doi.org/10.1038/s41586-024-07087-8
Promises and challenges
This study paves the way for the development of advanced therapeutics based on epigenetic silencing, offering a promising strategy to silence genes without altering their genomic sequence. The findings of this study hold immense potential for precision medicine and therapeutic interventions based on epigenome editing to target various diseases, such as hypercholesterolemia. However, the authors acknowledge the need for further validation in diverse models and potential challenges in clinical translation regarding safety and long-term efficacy.
Dr Cappelluti believes that an obstacle in expanding application of epigenetic editing to other organs is represented by the delivery. This is because, although LNPs can be used to deliver editors to the liver, many organs cannot be easily reached by LNPs. Moreover, LNPs are not amenable to a one-fits-all approach.
»The chemistry and parameters of the LNP formulation can affect silencing efficiency, and LNPs need to be optimised for the specific cargo. This is one of the challenges of working with LNPs,« said Dr Cappelluti. He added that, in addition to the delivery method, the editor design should also be optimised for the specific gene target.
Further work is needed to refine the delivery mechanisms, further enhance the specificity of editors, and expand the spectrum of target genes and therapeutic applications. Furthermore, further studies are required to determine the long-term effects of this approach.
»When we started working on this study, nobody believed in the possibility that epigenome editing would be applied in a clinically relevant setting, but our study demonstrates the feasibility of using epigenome editing to effectively silence genes that are therapeutic targets,« noted Dr Cappelluti. »As we continue to explore the power of the epigenome, we move one step closer to a future where diseases can be treated more effectively and safely than ever before.«
Link to the original article in Nature:
Durable and efficient gene silencing in vivo by hit-and-run epigenome editing
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







