Interview: Multiplex Base Editing Safely Enhances Sickle Therapy
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
»We have developed a precise base-editing approach for sickle cell disease that reactivates fetal haemoglobin, thereby preventing red blood cells from sickling. Unlike traditional CRISPR methods, our approach does not involve cutting the DNA, significantly reducing associated risks and offering a safer therapeutic option for patients,« the study's first author, Letizia Fontana, explains to CRISPR Medicine News. She is a postdoctoral researcher at the Université Paris Cité in France.
This study addresses a critical limitation in current sickle cell disease gene therapy – the variable and often insufficient levels of fetal haemoglobin (HbF) reactivation achieved through single-target CRISPR-Cas approaches. The FDA-approved therapy Casgevy, targeting the +58-kb BCL11A enhancer, shows promising but inconsistent results, with patients retaining substantial sickle haemoglobin levels and showing variability in the extent of HbF reactivation among individuals. The research team hypothesised that simultaneous editing of multiple enhancer regions could maximise BCL11A downregulation and achieve more consistent therapeutic outcomes.
The investigators employed CRISPR-Cas base editing technology to simultaneously disrupt GATA1 binding sites in the +58-kb enhancer and ATF4 binding sites in the +55-kb enhancer of BCL11A (see Figure 1). Unlike conventional nuclease approaches that create double-strand breaks, base editors introduce precise single-nucleotide changes, converting cytosines to thymines or adenines to guanines without generating potentially harmful chromosomal breaks.
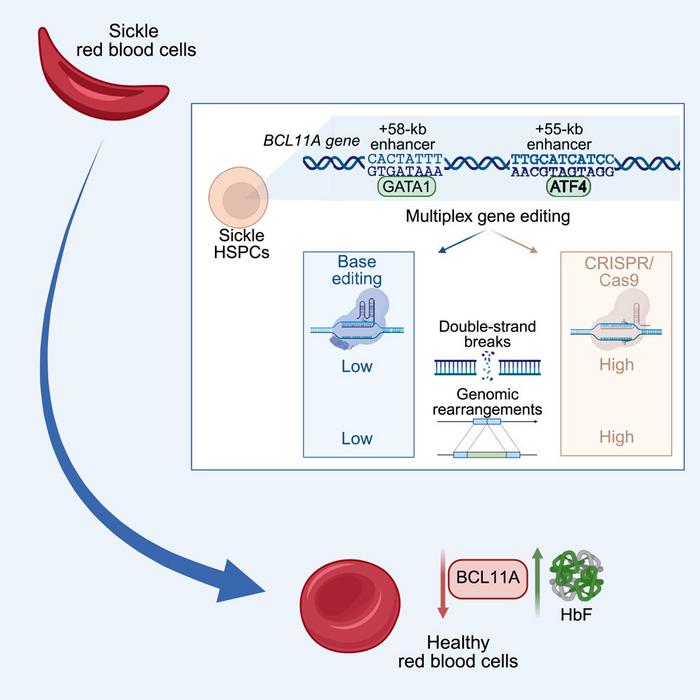
Through systematic screening of different base editor combinations in sickle cell disease patient-derived haematopoietic stem and progenitor cells, the researchers identified optimal editing profiles for each target site. The +58 CBEI profile (targeting the GATA1 motif) and +55 CBEII profile (targeting the ATF4 motif) emerged as the most effective single-target approaches.
However, the key finding was that simultaneous targeting of both enhancers produced superior results to either approach alone, with multiplex base editing correlating with stronger HbF reactivation compared to single editing. The multiplex approach achieved HbF levels of approximately 29% of total haemoglobin – sufficient to provide therapeutic benefit for sickle cell disease patients (see Figure 2).
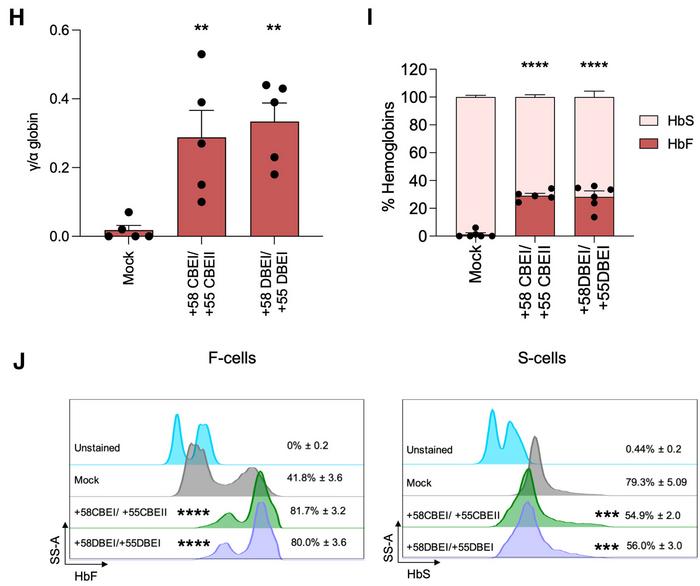
»By fine-tuning the expression of the BCL11A gene, which encodes a major transcriptional repressor of fetal haemoglobin, we effectively reactivated fetal haemoglobin to levels exceeding those achieved with the Casgevy strategy. Our results demonstrate a safe and durable approach for treating sickle cell disease,« Letizia Fontana explains.
Importantly, multiplex Cas9 editing at the same sites resulted in frequent 3.2-kb deletions or inversions, detected in one-third to nearly half of the treated cells. In contrast, base editing generated virtually none of these large genomic rearrangements.
In vivo xenotransplantation studies in immunodeficient mice demonstrated that the multiplex base editing approach successfully targets long-term repopulating haematopoietic stem cells. Edited cells maintained high editing efficiency and robust HbF expression 16–17 weeks post-transplantation, indicating the durability of the therapeutic effect. The researchers noted that "multiplex base editing does not affect engraftment and multilineage differentiation of HSCs", confirming the approach's compatibility with stem cell function (see Figure 3).
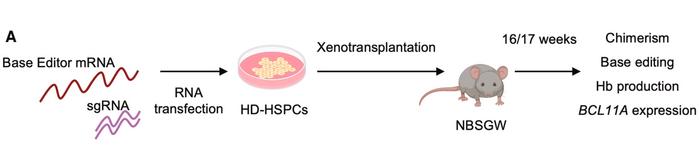
Comprehensive safety analyses, including whole-exome sequencing, RNA sequencing, and GUIDE-seq off-target detection, revealed minimal genotoxicity. Base editors showed significantly fewer off-target effects than CRISPR-Cas nucleases, with most detected off-targets occurring in non-coding genomic regions. Importantly, no insertions or deletions were detected at off-target sites, suggesting that base editors minimise the possibility of generating double-strand break-induced genomic rearrangements.
The study employed multiple complementary techniques to assess therapeutic efficacy, including reverse-phase high-performance liquid chromatography for globin chain analysis, cation-exchange HPLC for haemoglobin tetramers, and functional sickling assays under hypoxic conditions. These analyses confirmed that the frequencies of sickle cells in edited samples were reduced to levels similar to those observed in heterozygous, asymptomatic carriers of sickle cell disease. Yet, the authors acknowledged that phenotypic rescue was incomplete and variable across donors, underlining the need for further optimisation.
»Using base editors, we precisely disrupted regulatory elements in the BCL11A gene without inducing double-strand breaks, thereby reactivating fetal haemoglobin to therapeutic levels. This approach avoids genomic rearrangements while preserving long-term hematopoietic stem cell function,« Letizia Fontana adds.
This work demonstrates that multiplex base editing offers a safer and more effective approach to targeting BCL11A for sickle cell disease treatment, potentially addressing current limitations in clinical gene therapy approaches while providing a universal therapeutic strategy for both sickle cell disease and β-thalassemia patients.
The study was led by Panagiotis Antoniou and Annarita Miccio at Université Paris Cité (Imagine Institute) and conducted in cooperation with researchers from several institutions in France and Italy. It was published in Cell Reports Medicine on 21 October 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleCMN HighlightsNewsBeta ThalassemiaSickle Cell Disease, SCDHaemoglobinopathiesBase editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







