New Study Explains Why gRNA Efficiency Varies at On- and Off Target Sites
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Next CMN Webinar 29th June 3:00 PM CEST
Title: Multiplexed Genome Regulation In Vivo with Hyper-efficient Cas12a
Speaker: Dr. Lucie Guo,
Registration here

The applications of precise genome engineering using CRISPR in biotechnology and medicine are innumerable. Increasing in vitro, pre-clinical, and clinical data highlight CRISPR as a powerful tool for the treatment of serious diseases with a genetic basis, including cancers, cystic fibrosis, sickle cell disease, and atherosclerosis. However, adverse events due to off-target cleavage remain a significant health concern that limits the translation of CRISPR into new therapies.
One major drawback of CRISPR-Cas9-mediated genome editing is that not all guide RNAs (gRNAs) cleave target DNA efficiently. While the heterogeneity of gRNA activity is well recognised, exactly how CRISPR-Cas9 activity is regulated is not fully understood.
In a recent study, a team of researchers at the Center for non-coding RNA in Technology and Health (RTH), Department of Veterinary and Animal Sciences, University of Copenhagen, the Department of Biomedicine, Aarhus University, and the Lars Bolund Institute of Regenerative Medicine, and BGI-Research (China), used an energy-based model to identify mechanisms regulating CRISPR-Cas9 activity and specificity. They found that Cas9 "sliding" or binding competition on overlapping protospacer adjacent motifs (PAMs) affected gRNA activity by regulating local gRNA-DNA interactions.
»In this study, we identified a role for binding free energy changes in gRNAs efficiency at on- and off-target sites,« said Jan Gorodkin, Professor at RTH and the Department of Veterinary and Animal Sciences at the University of Copenhagen. »This sweet spot of binding free energy quantitatively defines the optimal activity of CRISPR-Cas9 at on- and off-target sites in that the gRNA is not binding too weakly or too strongly,« he added. Dr Gorodkin oversaw the computational work undertaken in this study.
»Our study provides further insight into why CRISPR-Cas9 can cleave off-target sites with higher efficiency than on-target sequences,« said Giulia Corsi, postdoctoral researcher at RTH and first author of the study. “By taking into account the role of the PAM context and local Cas9 sliding on overlapping PAM sequences, we can design more specific gRNAs, thereby increasing the efficiency and specificity of CRISPR-Cas9-mediated genome editing.«
The findings of this study were published today in Nature Communications.
Key unanswered question: How do the properties of gRNAs regulate CRISPR-Cas9 activity?
gRNAs regulate the activity and specificity of CRISPR-Cas9 by binding to a complementary target DNA sequence upstream of a PAM sequence. For Streptococcus pyogenes Cas9 (SpCas9), the 20-nucleotide sequence at the 5' end of the gRNA determines the specificity of the cleavage, and the canonical PAM sequence is 5'-NGG- 3'. Binding of gRNA to the target sequence upstream of PAM and the formation of a gRNA-DNA heteroduplex leads to the activation of Cas9, which introduces double-strand breaks (DSBs) at the target DNA.
The efficiency and specificity of the Cas9-induced DSBs vary depending on multiple factors. Low efficiency Cas9-mediated cleavage can hinder the efficacy of CRISPR-mediated genome editing, while low on-target specificity can be a result of competing off-target cleavage.
»It is becoming increasingly evident that the sequence and structure of the gRNA influence the efficiency and specificity of Cas9-mediated cleavage of target DNA sequences,« said Dr Yonglun Luo, who then continued: »However, exactly how the properties of gRNAs regulate CRISPR-Cas9 activity is not fully understood.” Dr Luo is an Associate Professor at the Department of Biomedicine, Aarhus University, and he oversaw the experimental work undertaken in this study.
Approach: Using an energy-based model to study CRISPR-Cas9 efficiency and specificity
To comprehensively investigate how the properties of gRNAs regulate CRISPR-Cas9 activity, the team used their previously developed energy-based model to analyse the role of gRNA binding patterns and changes in binding free energy and unfolding free energy in Cas9 activity.
»A first version of the binding free energy model was conceived in 2017 for the prediction of off-target sequences for RNA silencing and it was subsequently used as outset for the CRISPR-Cas9 binding free energy model,« said Dr Gorodkin. The team were the first to publish this model to predict CRISPR-Cas9 off-targets and the on-target specificity, which you can read more about in a previous CMN interview.
Dr Gorodkin explained that in addition to investigating the mechanisms underlying Cas9-gRNA binding to target sequences, their model enabled the study of Cas9 "sliding" between overlapping and thereby competing PAMs.
gRNA/DNA binding free energy changes affect the on-target cleavage activity of Cas9
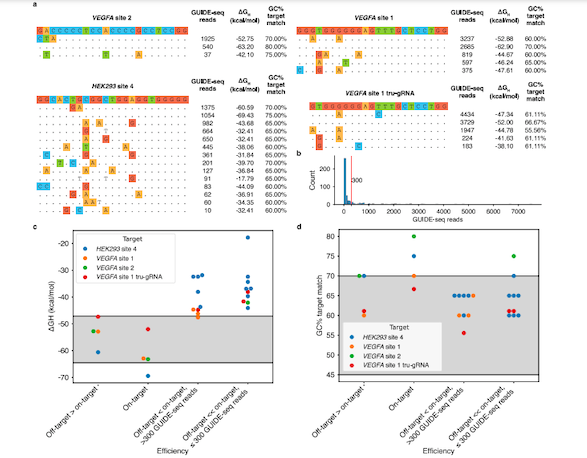
The team investigated the binding free energy, ΔGH, of the gRNA-DNA interactions and the effects of the energy changes upon gRNA-DNA hybridisation on the efficiency of 11,602 experimentally validated Cas9 gRNAs in other studies including a study published by the team in May 2021. The efficiency of gRNAs was measured as indel frequency, which is the most accurate indicator of CRISPR-Cas9 activity.
They found that highly efficient gRNAs provided a relatively narrow binding free energy range, which excludes extremely weak and strong bindings. Increasing gRNA GC content is one of the most robust ways to optimise CRISPR-Cas9 cleavage efficiency.
Interestingly, the energy-based model showed that hybridisation free energy change played a more accurate role in cleavage efficiency than GC content. In fact, increasing the GC content led to gRNA bindings that were too strong, and resulted in weak cleavage efficiency (Figure 1).
The team also found that some off-targets with high affinity to the preferential ΔGH range are cleaved more efficiently than other sites, including the on-target site, suggesting that ΔGH is a suitable indicator of cleavage activity. A similar distinction could not be achieved using the GC content alone.
Additionally, the team confirmed previous findings that gRNAs with stable self-folding structures result in low cleavage activities. A scaffold sequence is attached to the gRNA to facilitate the binding of Cas9 to the gRNA. Scaffold sequences can interact with bases in the gRNA, which can disrupt the correct folding of the scaffold.
The team found that highly efficient gRNAs had 3' seed regions that form strong interactions (lower free energy change) with the DNA. The 3' seed end of highly efficient gRNAs favour guanine at N19-N20 and cytosine at N18-N19. »Our findings highlight the important role of energetically favourable binding and folding interactions between gRNA and DNA, which promote Cas9 cleavage activation,« noted Dr Corsi.
Local Cas9 sliding influences gRNA efficiency and specificity
The presence of adjacent alternative overlapping PAMs upstream or downstream of the on-site PAM can make Cas9 compete for these and “slide”, resulting in either increased or decreased efficiency, respectively.
The team assessed this phenomenon by adapting their energy-based model accordingly, which resulted in a better separation of gRNAs with low, moderate, and high efficiency. »The model allows for more accurate calculation of the overall specificity of the gRNA when targeting local competing PAMs,« Dr Gorodkin added.
Experimental validation of the importance of local Cas9 sliding
To validate the effect of on-target PAM context on gRNA efficiency, the team measured CRISPR-Cas9 gene editing efficiency at more than 1,000 sites within four genes (ADRA2C, BMP3, CHRNB2, and PAX5) in human embryonic kidney (HEK293T) cells that were transduced with lentiviruses expressing gRNAs. gRNA efficiency was measured as indel frequency by deep sequencing. The experimental results validated the effect of local Cas9 sliding on gRNA efficiency and specificity.
Sites with an upstream PAM showed an 11.31% increase in mean efficiency, whereas sites with a downstream PAM exhibited a 12.13% decrease in mean efficiency, compared to sites with no alternative PAM as context. Notably, overexpression of Cas9 in HEK293T cells increased the cleavage efficiency at sites flanked by non-canonical PAMs.
Sliding affects PAM recognition by Cas9 variants
The team also studied sliding for overlapping PAMs in six SpCas9 variants with improved specificity and/or altered PAM compatibility. A preference for up- and downstream sliding towards canonical PAMs was observed for wild-type and certain high-fidelity variants. As expected, variants with increased specificity showed lower tolerance for sliding as this results in bulges between the gRNA-DNA binding and the Cas9 PAM binding site.
Going forward, the team will continue to improve its methods, which will include further optimisation of gRNA design for greater efficiency and specificity. »We are also interested in finding new ways to identify on- and off-target sites in a high-throughput manner, as well as developing innovative ways to address the off-target issues of CRISPR-Cas9, which is a key factor limiting its clinical translation.« Dr Luo said.
Link to the original article in Nature Communications:
CRISPR-Cas9 gRNA activity depends on free energy changes and on the target PAM context
Christos Evangelou, Ph.D., is a freelance medical writer and science communications consultant.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.