Prime editing is the new versatile 'word processor' for genome editing. Interview: Andrew Anzalone
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
(Interview is condensed and edited for clarity)

- congratulations on your paper being named top 10 of 2019 by Nature, what do You think of that?
Oh yeah, that was very exciting. I was not expecting all this, but it's been a lot of fun, and I am just happy to see other people are as excited about it as we are.
- Please, tell me a bit about your background and what led to the paper?
Well, with regard to my background, I am interested in human health and the development of therapeutics, and I did a medical degree and a Ph.D. in a combined program. My scientific background is in chemistry and molecular biology. During my graduate work, the whole explosion of CRISPR Cas9 genome editing was happening, which was very exciting to me.
However, for all the wonderful qualities of the CRISPR Cas nuclease, in and of itself, it can't really be used to correct most genetic diseases. The Cas nuclease itself makes double-strand breaks in DNA very efficiently, creating insertion and deletion mutations (indels) that are great for disrupting a gene–this is used extensively in research. But I think most people would agree that the best thing you could do is to correct the problematic mutation directly.
- Right?
And to do that with CRISPR nucleases is through a process called homologous recombination or homology-directed repair. That process uses a Cas nuclease to create a double-strand break and a donor DNA template that the cell will use to direct the repair process and correct the mutation. But homologous recombination is a generally inefficient process even in the best-case scenarios, and you usually end up with a minority of products being the intended edit and the majority being products that contain undesired indels. There are exceptions like ex vivo applications of blood stem cells, but in many other cases - especially in vivo editing - it's not currently seen as a good option.
Andrew Anzalone
Andrew Anzalone, Ph.D., MD, is a postdoc in the Liu Group at the Broad Institute of MIT and Harvard University, US, where he is developing the prime editing technology to make it more efficient as a tool for research and ultimately gene therapy. He did his Ph.D. at Columbia University, New York in the lab of Virginia Cornish, who was a graduate student alongside David Liu in the lab of professor Peter Schultz at Berkeley (now Scripps).
Using RNA instead of DNA
- That's where prime editing enters the scene?
Well yes, what is astonishing about CRISPR Cas nucleases is that they can find a particular place in the genome and make a cut given that there are three billion other base pairs around.
That's a wonderful property, but how can we adapt that system to achieve what we really want, which is to install a very precise edit?
So I thought, why not do that with RNA.
Fundamentally what's the difference?
DNA and RNA are both nucleic acids, and they both code the same information. The challenge is you need to be able to transfer that information from that RNA to genomic DNA.
- Right and use a reverse transcriptase like in PCR?
Yes. And using RNA presents a few advantages - the CRISPR Cas system already uses RNA for the targeting. A spacer sequence and a CRISPR guide RNA with a nucleotide sequence, that specifies where Cas9 will bind. And so, in theory, why not make an RNA that both specifies where Cas9 will bind and also determines what your edit will be?
And right around the time, I started considering postdocs, David Liu's lab came out with base editors. The concept of base editors is you fuse an enzyme that modifies the bases to an impaired version of Cas9 that only nicks one of the two DNA strands. Depending on the base editor variant, you can make a C to T change or an A to G change, and that lets you do a lot actually. But still for all their good qualities base editor can only do 2 out of 6 types of point mutations and they can't do deletions or insertions.
- Ok, but if you fuse the reverse transcriptase to Cas9, you might?
Yes. That was the idea, but how do we make this happen?
So, when Cas9 nicks one strand of the DNA, that strand becomes very flexible and could, in theory, be used as a primer for reverse transcription of a new strand of DNA. The guide RNA has been engineered to be a more sophisticated guide RNA. It still has the spacer sequence that defines the target, but now it also has this extra sequence that's used as a template for the reverse transcriptase. We call that a prime editing guide RNA or a pegRNA.
When the nicked strand has a 3' end where new individual bases can be added on - then, in theory, you can write new information on the genomic DNA directly.
So compared to homology-directed repair, where the repair machinery and template have to come in, the prime editing process is carried out by the molecular machine itself, directly installing an edited piece of DNA onto the genomic DNA.
[See infographic]
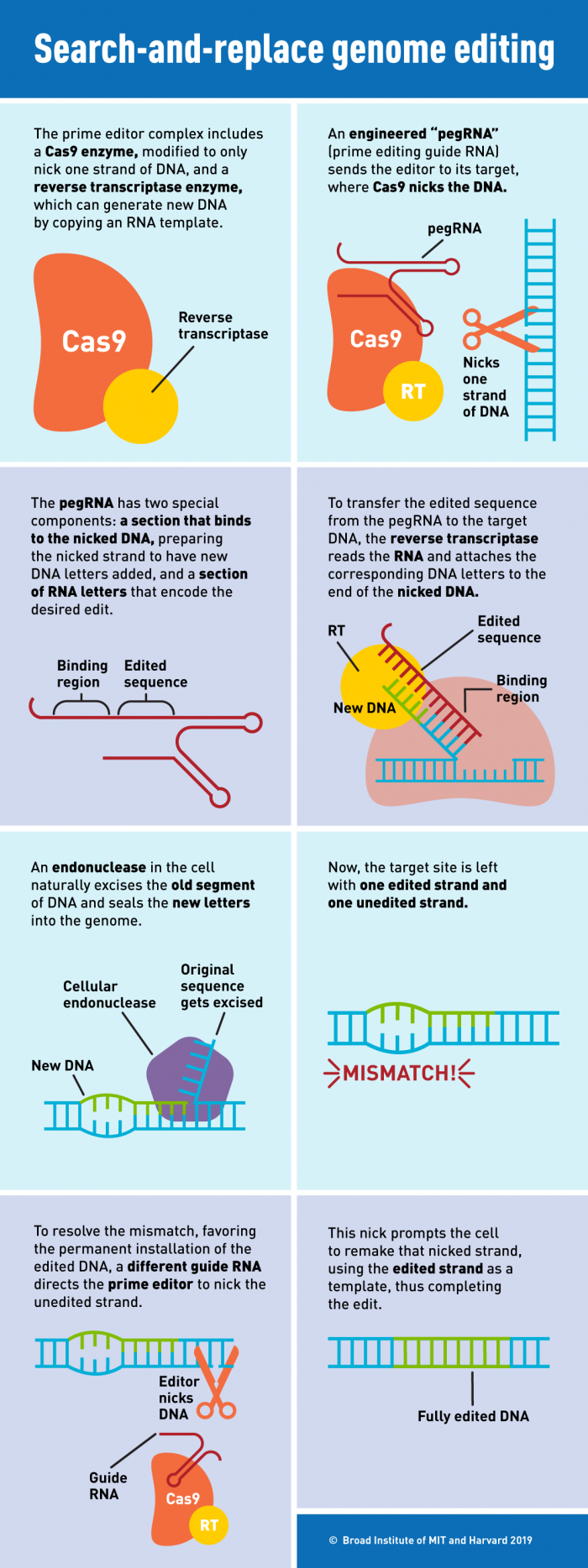
Point mutations, deletions, and insertions
- and how did it eventually work?
We were able to show that you can make any type of point mutation, make insertions up to about 40 nucleotides, make deletions up to around 80 nucleotides, and one of the significant features is that you can make point mutations pretty far away from the cut site.
We're able to make a point mutation up to 31 nucleotides away from where the nick occurs. This contrasts with homology-directed repair, where you usually have a pretty short window of editing around eight base pairs from where the cut happens - and it's in principle the same with base editors.
So with prime editing you can edit positions that are further from the cut site, and the PAM requirement of Cas9 is a bit lower.
- what about off-target effects?
We have fewer off-target effects with prime editors compared to Cas9. We did a study where we used previously characterized Cas9 off-target sites and then treated them with the prime editors. We did deep sequencing of those off-target places, and prime editors really don't generate a lot of off-target edits. But that doesn't mean there are no off-targets elsewhere in the genome. We can't know that without essentially doing whole genome sequencing on single cell derived clones, which is very costly.
I think off-targets is a consideration in the entire field of genome editing and gene therapy, and all of those challenges that come with other forms of gene editing also come with prime editing.
Size and delivery are challenges
- What do you think are the main limitations are?
Delivery is one of the challenges with prime editing. The standard Cas9 protein is coded for by about 4.1 kb of DNA, and the prime editor protein requires about 6.3 kb in its current version, so it's quite a bit larger, and that just makes it a little bit more difficult to pack into a virus (currently one of the more appealing delivery methods). We were able to design a system that split the editor proteins into two pieces, and when two viruses co-infect the same cell, they can reconstitute the editor protein and carry out editing.
But I think another promising delivery option is mRNA encapsulated in lipid nanoparticles because it doesn't have the same fundamental size restriction that viruses tend to have. I think that's an excellent option for therapeutics.
- What are you working on now?
With other members of the Liu lab, one of our efforts is to compress the components required so we could fit it all in an AAV (Adeno-Associated Virus), which can carry around 4.5 to 5 kb.
We also want to identify the rate-limiting steps in prime editing and try to determine if there's anything we can do to make it more efficient, such as engineering the reverse transcriptase enzyme to be more active. And we rely a lot on the DNA repair in the cells, so if we can figure out what those important DNA repair proteins are, we may be able to steer the repair process toward our desired edit.
And then, all of the in vivo applications. I think prime editors could be great as a research tool already. Still, the ultimate goal is to make this therapeutically relevant, so the Liu Lab is working on addressing the challenges associated with that.
- excellent, thank you very much.
Search-and-replace genome editing without double-strand breaks or donor DNA
The study 'Search-and-replace genome editing without double-strand breaks or donor DNA' was published in Nature by Andrew Anzalone, MD, PhD, and professor David Liu, and a team from the Broad Institute of MIT and Harvard University.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







