Toward Safer Genetic Engineering, One Base at a Time
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
First developed in Dr. David Liu’s lab at Broad Institute in 2016, base editors are engineered as fusion enzymes that can specifically modify a single cytidine or adenosine base in genomic DNA.
A base editor consists of a Cas9 variant whose RuvC domain (the nuclease domain that initiates cleavage of the non-complementary DNA strand) has been inactivated, and one of two specific DNA modifiers that can alter single bases. Mutational deactivation of the RuvC domain in Cas9 results in a nuclease that can only cleave – or nick – one of the two DNA strands, and this is referred to as nickase Cas9 ( nCas9).
Two classes of base editors
Base editors can be distinguished as either cytosine or adenosine base editors. These differ in the type of DNA-modifying enzyme that is fused to nCas9.
Cytosine base editors (CBEs) comprise nCas9 fused to a cytidine deaminase enzyme that can convert cytidine to uracil, both of which have similar base-pairing characteristics.
Adenosine base editors (ABEs) comprise an engineered ssDNA-specific adenosine deaminase, which converts adenosine to inosine, which can be read as guanine by the DNA replication and transcription machinery.
How base editing works
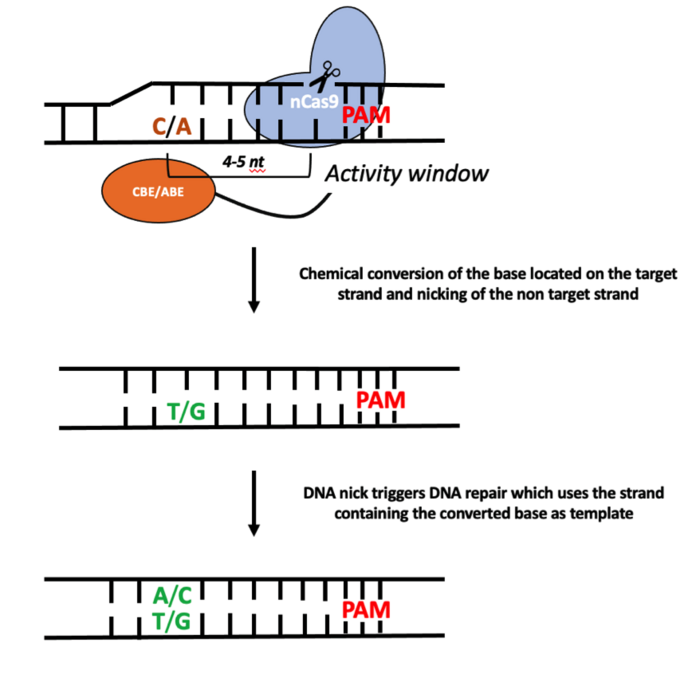
Although structurally different, CBEs (C-G to T-A) and ABEs (A-T to G-C) share a common mode of action; the CBEs and ABEs chemically modify the non-target strand while the base on the opposite strand is nicked by the nCas9. Base editors are guided toward the intended target in the same way as a standard CRISPR-Cas9 endonuclease. This means that the 20-nucleotide (nt) spacer sequence of the single-guide RNA (sgRNA) will define the target (see Figure 1).
A suitable protospacer-adjacent motif (PAM) must also be located immediately next to the sgRNA target. The exact PAM requirement will depend on the Cas9 species used. For example, the nCas9 that is derived from the standard spCas9 requires NGG as the PAM. Subsequently, the single strand DNA repair pathway uses the newly converted base (C to T or A to G) as a template to repair the nick introduced by the nCas9.
It is important to note that base editors possess a so-called “activity window”, which is the area within which bases are converted. The activity window for CBEs is generally 5 nucleotides, and for ABEs this is 4 nucleotides. This means that – for example in the case of CBEs – all the C(s) present within 5 nucleotides from the CBEs binding site can be potentially converted to T. Conversion of unintended bases is known as bystander editing.
The conversions are stable
One obvious question is why the cellular DNA repair machinery does not try to reverse the conversions carried out by base editors, given that a chemical alteration to genomic DNA is not as invasive as a double-stranded break (DSB).
Among the many cellular repair pathways is base excision repair (BER), whose function is to remove and correct erroneously incorporated bases from the genome throughout the cell cycle. Theoretically, this should impair the efficiency of a CBE. To counteract this risk, researchers have equipped the CBEs with a uracil glycosylase inhibitor (UGI), which greatly increases the rate of C to T conversion in living cells. Such a strategy is not required for ABEs, possible because adenosine to inosine conversions are relatively rare in nature, thus the need to evolve a dedicated countermeasure was minimal.
Bypassing the need for double-stranded breaks
The advent of genome editing creates the possibility to address genetic diseases at their very root, the DNA. The introduction of a DSB is an inevitable step when standard designer nucleases (DNs) such as CRISPR-Cas9 are employed. This triggers and exploits certain DNA repair mechanisms such as the non-homologous end joining (NHJ) and homology-directed repair (HDR) pathways, which cells have evolved as a defense against DSBs. This process makes it possible to inactivate genes, disrupt regulatory regions, or to precisely edit sequences by adding new information (targeted integration) or modifying the one already present (single nucleotide changes).
While the aforementioned strategies are being explored in ongoing clinical trials, concerns exist regarding the safety of deliberately introducing a DSB as part of a gene-editing procedure. These concerns are substantiated by reports highlighting how chromosomal aberrations and undesired small and large-scale modifications could be a possible byproduct of gene-editing interventions. As consequence, base editing has two great advantages over other gene-editing modalities: i) no DSB is introduced, and ii) since no DSB is introduced the editing outcome won’t depend on the engagement of the HDR pathway, whose DSB repair frequencies are known to be lower than the imprecise NHEJ pathway.
Clinical applications of base editing: single base changes for a better outcome
The possibility to change one base at a time might seem like a modest development in the genome-editing field. However, the clinical potential of base editing is highlighted by the fact that of the >50,000 mutations (e.g. deletions, insertions) known to be associated with human genetic conditions, 33,739 are actually point mutations.
Of these, 47.1% could theoretically be corrected by performing an A-T to G-C (ABEs) and 13,5 % a C-G to T-A conversion (CBEs), thus providing a possible solution for more than 60% of the genetic conditions caused by a single-nucleotide variant (SNV).
Within research related to human disease, some of the most compelling applications of base editing so far are the restoration of functional laminA in a Progeria mouse model using ABEs, and the rescue of dystrophin expression in mice using base editors to skip exon 50, thus leading to partially functional dystrophin that may slow disease progression.
The clinical potential of base editing is further highlighted by Verve Therapeutics, which recently announced robust pre-clinical data for its base-editing candidate VERVE-101, an adenosine/cytidine base editor designed to disrupt the PCSK9 protein by creating a premature stop codon and thus lower blood levels of low-density lipoprotein cholesterol (LDL-C), as a treatment for atherosclerotic cardiovascular disease (ASCVD).
Other examples include the BEAM-101 and BEAM-102 programmes by Beam Therapeutics for the treatment of sickle cell disease (SCD). These are designed to either create a point mutation that results in increased foetal haemoglobin levels (BEAM 101) or to convert the SCD mutation into the Makassar variant (BEAM-102), which is a normal human variant of the major SCD-associated gene.
Base editing future development
Base editing represents an alternative approach to other designer nucleases, to precisely correct clinically-relevant point mutations without the risks associated with DSBs. Its therapeutic potential is highlighted by the aforementioned therapy development programmes. Constant improvements to base editors should minimise by-stander editing by engineering base editors variants with a narrower editing window, and the target range should be expanded by engineering base editors with reduced PAM stringency.
Base editors are also being explored in explorative studies as possible treatments for Alzheimer’s disease, and the debilitating and fatal disorder Amyotrophic lateral sclerosis (ALS). These advances indicate how base editors may redefine and direct the future of precision medicine toward a safer approach where the precise change of just one letter may dramatically ameliorate severe genetic conditions that are currently incurable.
Related CMN Explainers:
Prime Editing: A Double-Strand Break Free Solution for Safe and Efficient Gene Editing.
Antonio Carusillo, PhD, currently employed at Vector BioPharma.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.