Breaking: CRISPR screen identifies endosomal trafficking modulators
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Using an engineered splice-switching reporter system, scientists demonstrated that AP1M1 knockout dramatically enhances ASO activity by delaying endosome-to-lysosome transport both in vitro and in vivo. This delay provides ASOs with extended residence time in endosomal compartments, increasing their probability of cytoplasmic escape. Conversely, TBC1D23 knockout reduces ASO efficacy by disrupting AP-1 vesicle tethering at the trans-Golgi network.
“We performed an unbiased CRISPR/Cas9 knockout screen with a genetic splice reporter to identify genes that can increase or decrease ASO activity, resulting in the most comprehensive catalog of ASO-activity modifier genes”Malong et al.
The researchers developed a sophisticated fluorescent reporter to quantify ASO nuclear activity in real time (see Figure 1). The system employed a modified EGFP gene interrupted by a β-globin intron-2 sequence containing specific mutations that create a cryptic splice site. Under normal conditions, this aberrant splicing prevents functional EGFP expression. However, when a splice-switching ASO successfully reaches the nucleus and binds to its complementary sequence within the cryptic exon, it redirects splicing to produce correctly spliced, functional EGFP. This provides a direct functional readout of nuclear ASO activity.
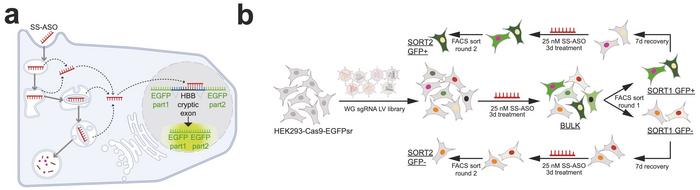
The whole-genome CRISPR knockout screen utilised a comprehensive sgRNA library targeting approximately 18,000 genes with multiple guide RNAs per gene, totalling over 75,000 individual sgRNAs. Following puromycin selection and treatment with 25 nM splice-switching ASO for 72 hours, cells underwent fluorescence-activated cell sorting to isolate the top and bottom 10% of GFP-expressing populations. This process was repeated in two consecutive rounds with recovery periods between treatments to enrich for genuine modulators of ASO activity.
»We performed an unbiased CRISPR/Cas9 knockout screen with a genetic splice reporter to identify genes that can increase or decrease ASO activity, resulting in the most comprehensive catalog of ASO-activity modifier genes,« the authors state in their Nature Communications paper.
The screen identified distinct categories of ASO activity modulators with clear functional themes. Genes that enhanced ASO activity upon knockout showed significant enrichment for clathrin and cargo adapter activity, particularly components of the AP-1 clathrin adapter complex (AP1M1, AP1G1) and coatomer proteins (COPG1, COPB1, COPZ1). Gene ontology analysis revealed that positive modulators were predominantly involved in Golgi-associated vesicle transport, trans-Golgi network function, and endosomal trafficking pathways.
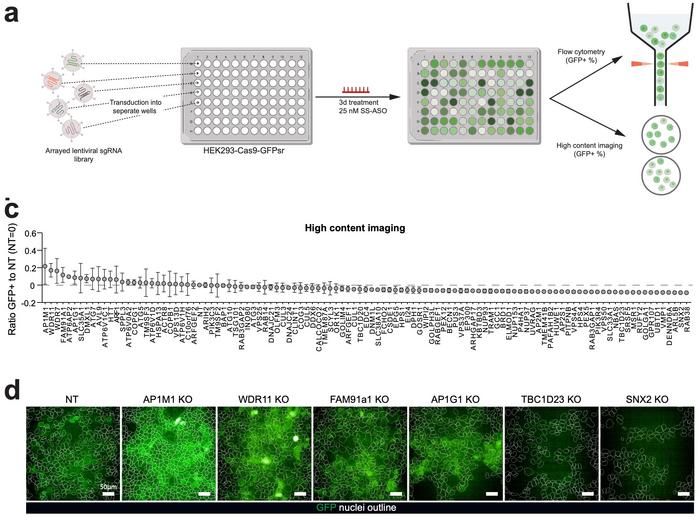
Validation through arrayed screening of 98 target genes confirmed these findings using individual sgRNAs and high-content imaging analysis (see Figure 2). AP1M1 emerged as the most potent enhancer of ASO activity, while TBC1D23 and SNX2 significantly reduced ASO efficacy upon knockout. Importantly, these effects were specific to the unassisted, passive uptake ASO uptake and did not affect lipofectamine-mediated transfection, indicating that the identified factors modulate post-uptake trafficking rather than cellular entry.
“We conclude that AP1M1 KO cells have
functional lysosomes and suggest that the increased activity of ASOs
may be due to an effect earlier in the traf cking pathway, speci cally
through more ef cient endosomal escape”Malong et al.
Detailed mechanistic studies revealed that AP1M1 knockout cells exhibited delayed transport of endocytic cargo to lysosomes, as demonstrated using DQ-red BSA tracer experiments. TGN46 protein levels were significantly reduced in AP1M1 knockout cells, indicating a disruption in retrograde trafficking from endosomes to the trans-Golgi network. The researchers successfully validated their findings in vivo using transgenic mice expressing the same splice-switching reporter, demonstrating a significant correlation between Ap1m1 knockdown levels and enhanced ASO activity in liver tissue.
The findings suggest that manipulating endolysosomal pathways could enhance therapeutic ASO delivery by prolonging endosomal retention before lysosomal degradation. As the authors put it:
»We conclude that AP1M1 KO cells have functional lysosomes and suggest that the increased activity of ASOs may be due to an effect earlier in the trafficking pathway, specifically through more efficient endosomal escape.«
The research was conducted by teams from F. Hoffmann-La Roche, Genentech, Roche Innovation Center Copenhagen, and University of Basel. It was published in Nature Communications on 30 June 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleBreaking newsCMN BriefsCMN HighlightsNewsCRISPR ScreensRoche
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.