CATCHing Tumour DNA: Researchers Develop Engineered Bacteria to Detect Cancer in the Gut
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Despite advances in cancer treatment, colorectal cancer (CRC) remains a significant cause of morbidity and mortality worldwide. Although early cancer detection can improve outcomes, current non-invasive tests, such as faecal immunochemical testing, show limited accuracy. While effective, invasive diagnostic procedures, such as colonoscopy, are unsuitable for large-scale screening because of their high cost and risk. In addition, screening programmes using colonoscopy are only available for individuals older than a certain age, usually 45 years.
In a recent study, researchers at the University of California San Diego, South Australia Health and Medical Research Institute, and Colonoscopy Clinic used CRISPR to engineer Acinetobacter baylyi, a non-pathogenic bacterium commonly found in soil, to detect specific DNA mutations shed by CRC tumours into the gut lumen.
The researchers have shown that bacteria take up tumour DNA through natural competence and integrate target sequences into their genome through homologous recombination. This prompts a detectable change in the bacteria, allowing sensitive and specific identification of CRC mutations.
This proof-of-concept study provides evidence that engineered non-pathogenic bacteria can be used as biosensors to non-invasively detect tumour DNA in the gut, paving the way for the development of novel early cancer detection methods for large-scale CRC screening.
“In this study, we proved that cell-free DNA released from tumours can be used as an effective signal for bioengineering, providing a new and important input for synthetic biology applications. Our findings confirmed that this approach can be used to diagnose disease within a complex environment such as, but not limited to, the colorectal lumen,” said Dr Daniel L. Worthley of Colonoscopy Clinic, who was one of the lead investigators of the study.
“Although further development is required, this approach could help us detect primary early-stage gastrointestinal tumours. In the future, depending on the bacterial species used and the mode of administration, its clinical use could be significantly more extensive than that, including metastatic cancers and infections,” he added.
The study was published in Science.
Transforming bacteria into biosensors for detection of tumour DNA
Advances in synthetic biology have enabled reprogramming of microbes into living diagnostics and therapeutics. For example, bacteria can be engineered to detect disease biomarkers and produce easily measurable outputs, such as coloured molecules, in stool or urine.
A key advantage of bacterial biosensors is their ability to directly integrate cues from the complex environment of the gut. In the gastrointestinal tract, cell-free DNA has a short half-life because of the activity of intestinal enzymes, such as intestinal deoxyribonuclease (DNase), limiting the ability to analyse faecal samples for tumour DNA. Engineered bacteria in the gut can capture cell-free DNA released from CRC tumours before its degradation.
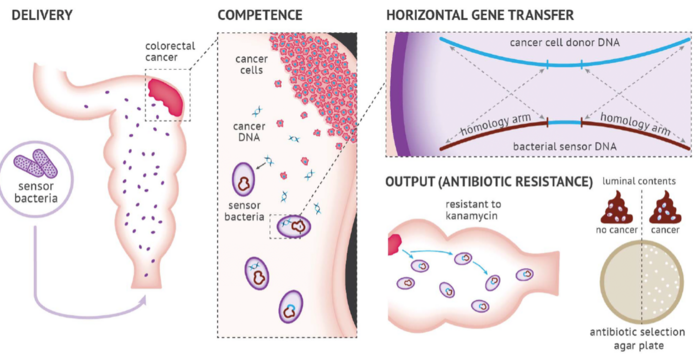
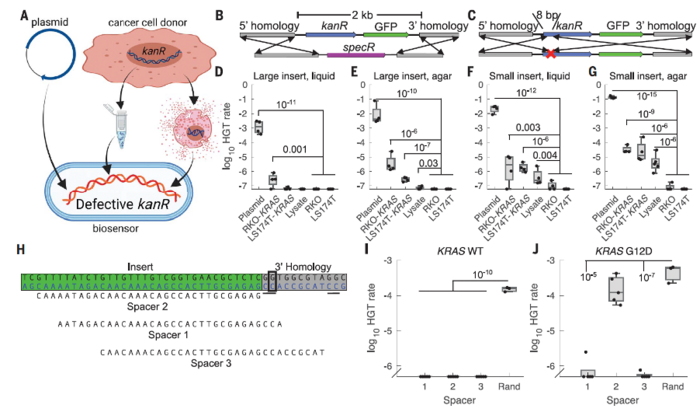
Researchers developed a method called ‘Cellular Assay for Targeted, CRISPR-discriminated Horizontal Gene Transfer’, or simply ‘CATCH’, to detect specific extracellular DNA sequences and mutations using bacterial biosensors. Specifically, they engineered non-pathogenic A. baylyi to detect donor DNA released from CRC cells, organoids, and tumours (Figure 1). Integration of tumour DNA into the bacterial genome confers resistance to the antibiotic kanamycin. As such, kanamycin selection can be used as an easily measurable output for detecting colorectal tumours.
Capitalising on the inherent ability of A. baylyi to take up foreign DNA through horizontal gene transfer (HGT), a phenomenon known as natural competence, this represents the first application of bacterial biosensors for targeted detection of cell-free DNA released from colorectal tumours.
Dr Worthley explained that the natural competence of Acinetobacter baylyi, coupled with the fact that it is a very well-studied, non-pathogenic organism that can colonise the colorectum, are reasons why they chose to engineer this bacterial species as a biosensor for the detection of tumour DNA in the gut.
Detecting cancer mutations in vitro
The authors confirmed that A. baylyi could take up and integrate tumour DNA containing an engineered antibiotic resistance gene cassette flanked by homology arms that matched the bacterial genome. Experiments using CRC lines demonstrated robust and specific detection of target DNA both in liquid culture and on agar without the need for purification.
CRISPR spacers were introduced into the bacteria to enable the detection of cancer mutations. Often, the sequences of mutant genes in cancer cells differ only slightly from those of wild-type genes. For example, in KRAS, an oncogene that is frequently mutated in CRC, a single guanine-to-adenine substitution is sufficient to produce a constitutively activated protein that promotes unrestrained cell growth.
The authors assessed the ability of the biosensors to determine the mutation status of KRAS in vitro using coculture assays. The sequence of the CRISPR spacers was designed in a way that they could determine whether the KRAS gene that was integrated into the bacteria was wild-type or KRASG12D.
“This was a superb idea by the co-first author of the paper, Dr Rob Cooper. He designed an RNA guide that would cut the normal wild-type KRAS gene, but not the mutant one,” Dr Worthley noted.
The authors tested CATCH using engineered tumour models expressing an antibiotic resistance gene flanked by sequences from the KRAS oncogene (Figure 2). Both CRC cell lines and 3D organoid models were engineered to release donor DNA. Biosensor bacteria took up and integrated tumour DNA, gaining antibiotic resistance. CRISPR spacers introduced into biosensors enabled single-base discrimination between mutant and wild-type KRAS. In this setup, only bacteria that had taken up mutant forms of KRAS could survive in the presence of antibiotics.
Commenting on the advantages of cellular testing over traditional laboratory-based methods for the detection of tumour DNA, Dr Worthley said, “A cellular biosensor can diagnose and treat diseases at the time and place of disease onset without requiring the processing and analysis of the sample ex vivo, which is time consuming and complex.”
He added that this approach is particularly useful for disease screening in settings where access to resource-rich laboratories and sophisticated assays is limited.
Detecting tumours in the gut
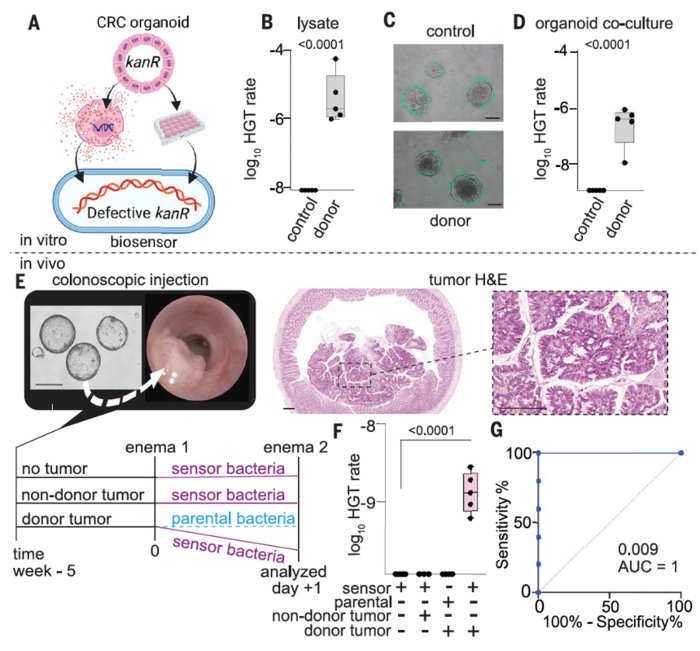
To validate the clinical relevance of their method, the team used CATCH to analyse the ability of sensor bacteria to detect KRAS mutations in CRC organoids and an orthotopic mouse model (Figure 3). When cocultured with organoid lysates or viable organoids, A. baylyi bacteria efficiently integrated donor sequences, with a log10 HGT rate of ~6 after 24 hours of coculture.
For the in vivo experiments, they administered the engineered A. baylyi bacteria rectally to mice harbouring colorectal tumours.
“This is not how we would apply this clinically, but this was a way that we could test large numbers of biosensors administered directly around the colorectal tumour,” Dr Worthley explained.
Rectal delivery of A. baylyi bacteria to mice with CRC donor tumours resulted in the development of kanamycin resistance in biosensor bacteria in luminal contents through HGT, with a transformation efficiency of 1.5 ×10−9. Notably, CATCH successfully discriminated between mice with and without CRC, providing an area under the curve (AUC) of 1 (P = 0.009).
The authors went one step further by designing an advanced biosensor to detect cancer mutations in non-engineered DNA to more closely resemble detection of tumour mutations in a patient. The system integrates a transcriptional repressor and reporter construct. Integrated mutant DNA displaces the repressor, triggering reporter gene expression. Spacer sequences carefully designed against a single nucleotide polymorphism ensure that only DNA with the cancer mutation, and not wild-type DNA, avoids CRISPR degradation and reporter activation.
CATCHing tumour DNA: promises and challenges
Although the authors acknowledge that further work is needed before clinical use, CATCH represents an important early step toward in situ mutation detection. This proof-of-concept study demonstrates that bacterial biosensors can capture and integrate target DNA in complex intestinal environments. CATCH could enable non-invasive detection of tumour DNA in the stool of patients with early-stage tumours that are more likely to be successfully treated.
Even though this study focused on KRAS mutations in CRC, the modular design of CATCH allows detection of any DNA sequence of interest by altering CRISPR spacers and homology arms. For example, HPV sequences could be used to detect cervical cancer, and BRAF mutations could be used to detect melanoma.
However, substantial optimisation is needed before clinical translation. Public acceptance of engineered organisms, albeit potentially beneficial, may pose challenges. Systems that prevent environmental release and limit horizontal gene transfer must be implemented.
In addition, future efforts are needed to increase the efficiency of tumour DNA detection and signal-to-noise ratio by detecting multiple mutations. Thorough evaluation of performance compared to standard diagnostics is critical for moving CATCH closer to the clinic. Future iterations may couple detection of tumour DNA with delivery of cancer therapeutics, such as anti-cancer peptides.
“Our next steps include improving the biosensors, finding ways to package them for oral delivery, and ensuring patient and environmental safety,” Dr Worthley noted. He added, “With continued development, engineered sensor bacteria may one day provide non-invasive diagnosis and personalised treatment of cancer and other diseases.”
Link to the original article in Science:
Engineered bacteria detect tumor DNA
For more information on the technology, visit www.catch.contact.
Christos Evangelou, PhD, is a freelance medical writer and science communications consultant.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.