CRISPR-Cas9 Causes Unintended On-Target Mutations in Early Human Embryos
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The mechanisms that govern cell fate in human embryos are the subject of investigations by Kathy Niakan’s group at the Francis Crick Institute, UK. The team uses CRISPR-Cas9 as a tool to disrupt and characterise various genes implicated in early human developmental processes and they recently made a surprising discovery.
After genome-editing of a set of early human embryos, they found large unintended mutations in 16% of the genomic DNA (gDNA) samples. These events included chromosomal and segmental copy number abnormalities, and they would have been missed by standard quality control tools normally used after genome editing.
Commenting on the significance of the findings, Eric Bennett, MSc. Dr. med., Scientific Advisor, COBO Technologies, who was not involved in this study, said:
»The concept of off-target analysis, which is currently addressed through short read NGS approaches, needs to be redefined to address the concerns raised in this work.«
Niakan’s group have now built an open-source pipeline based on next-generation sequencing (NGS) data that can be used to identify whether or not CRISPR-Cas9 has caused unintended on-target mutations. The results were published in PNAS last month, and we spoke with lead author, Gregorio Alanis-Lobato, to hear more about the work.
CRISPR-Cas9 as a tool to study early human embryogenesis
In 2017, the Niakan lab authored a study involving the use of CRISPR-Cas9 gene editing to characterise the role of the pluripotency transcription factor OCT4 during human embryogenesis. OCT4 is encoded by the Pou5f1 gene in humans, which is found on chromosome 6.
In that study published in 2017, CRISPR-Cas9 was deployed to disrupt Pou5f1 for research purposes, but groups elsewhere have investigated the possibility of using CRISPR-Cas editing to correct mutations in early human embryos as a means to reduce the burden of or even cure genetic diseases before birth.
Given recent findings about unintended CRISPR outcomes reported elsewhere, the Niakan lab sought to take a closer look at what happens to the genome in CRISPR-edited early human embryos, as Gregorio Alanis-Lobato explains:
»Other groups have recently shown complexity at on-target sites following CRISPR-Cas9 genome editing in mouse embryos, cell lines and primary cells. In light of these findings and our own previous work on OCT4, we thought it would be important to check for on-target complexity in early human embryos. The biological context was important to consider, as it was possible that early human embryos would respond differently to DNA double strand breaks induced by the CRISPR-Cas9 system compared to mouse embryos or cell lines.«
Revisiting the OCT-deficient CRISPR-edited early human embryos
During the previously published OCT4 study, the group CRISPR-edited zygotes that had been donated to research as surplus following in vitro fertilisation treatment.
Using single-cell amplified gDNA samples, they confirmed on-target genome editing in all microinjected embryos, with an expected indel pattern of mutations in most samples. However, they noted that genotype determination was not possible for a small number of samples because of failures to amplify the on-target gDNA region. At that time, the researchers attributed this to unsuccessful targeting events. However, as others began to report complex on-target editing events, the group wondered if their previous analysis methods had missed something.
Purpose-built computational pipeline to analyse embryonic gDNA
Given that conventional DNA analysis methods did not reveal complex on-target events in Pou5f1-edited embryos, the group developed a purpose-built computational pipeline, an undertaking that turned out to be challenging, as Alanis-Lobato explains:
»The most challenging aspect was that the genetic material we studied came from single cells or biopsies, i.e., clusters of roughly 5 cells, from early human embryos. Analysing data from such a small amount of material required the development of computational pipelines specifically adjusted to this type of data and included stringent preprocessing and filtering steps.«
The group succeeded in developing a set of bioinformatics tools that can analyse single-cell low-pass whole genome sequencing (WGS), transcriptome, and deep-amplicon sequencing data to specifically assess the prevalence of loss of heterozygosity (LOH) events in the genomes of CRISPR-Cas9-edited early human embryos.
Frequent loss of heterozygosity events in CRISPR-edited early human embryos
A LOH event occurs when either the maternal or paternal copy of a given gDNA region is lost, in this case after CRISPR-Cas9 genome editing.
The group’s computational pipeline searches sequencing datasets for single nucleotide polymorphisms (SNPs) in gDNA from the input samples. If one cell has heterozygous SNPs close to the CRISPR-Cas9 on-target site and another cell from the same embryo doesn’t have them, a LOH event is likely to have occurred.
The group reanalysed whole-genome amplification (WGA) data from 23 Pou5f1-targeted and eight Cas9 control samples. To mitigate any risk of data skewing with this sample size, the researchers microinjected an additional set of zygotes with either Pou5f1-targeting RNP or Cas9 alone and performed WGA and WGS on the resulting gDNA. After quality control checks, there was a total of 65 samples for analysis – 25 Pou5f1-targeted, 16 Cas9 controls, and 24 uninjected controls.
Upon analysing all samples, the group found that LOH events were more prevalent in Pou5f1-edited embryos compared to both Cas9-injected and Cas9-uninjected controls, with on-target LOH events in 32% of CRISPR-edited samples compared with 6.25% of control samples. These events encompassed chromosomal and segmental copy number abnormalities, and mirror other group’s findings of unintended on-target damage in somatic cells as well as mouse embryos.
Taken together, 16% of the samples exhibited gDNA losses or gains adjacent to the Pou5f1 locus and LOH events in the size range of 4-20 kb.
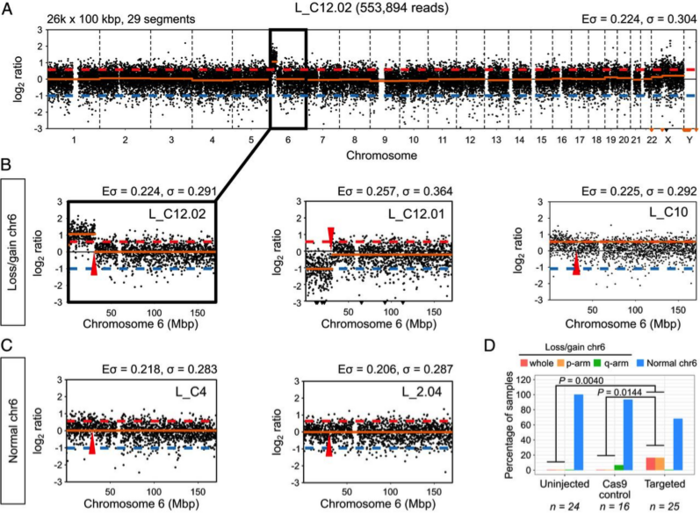
While the analyses undertaken in this work were based on events that occurred after Pou5f1 targeting, Alanis-Lobato points out that target site may impact the outcome:
»CRISPR-Cas9 genome editing fidelity could be locus-dependent. For example, there are studies that demonstrate that the proximity of the CRISPR-Cas9-targeted locus to the telomere significantly increases the possibility of inadvertent chromosome arm truncation.«
Surprising results that warrant increased vigilance in human genome-editing applications
Despite similar findings by others, albeit not in human embryos, the group was very surprised by its findings and points out the significance with respect to future clinical applications in embryos. Alanis-Lobato says:
»Editing-associated large deletions have been reported in early mouse embryos after the use of nickases and prime editing. We were very surprised to find that unexpected CRISPR-induced on-target mutations can occur in early human embryos. For research in human cells, especially where future clinical applications are concerned, we recommend to actively look for these unintended changes as they could have clinical implications for example in cancer.«
Eric Bennett echoes this recommendation:
»This work addresses a relevant concern about the specificity and efficacy of programmable nucleases in general. This and other recent studies that address the issue of unintended on-target outcomes all suggest that the conventional short read-based NGS approaches are not adequate to capture the full spectrum of outcomes induced by gene editing. The current study takes advantage of a deep amplicon long PCR-based approach, which also possesses limitations due to PCR bias, but the use of other methods such as the rapidly developing single molecule sequencing technologies such as PacBio and Nanopore sequencing will help to reveal the full spectrum of unintended on-target gene editing induced outcomes.«
The Niakan group continues to unravel the exact nature of the unintended outcomes observed at on-target sites, but CRISPR-Cas9 remains a valuable and reliable part of its toolbox:
»If we knew how early developmental processes work, our knowledge could improve the understanding and treatment of infertility and developmental disorders. We will continue to use CRISPR-Cas9 genome editing, but in full awareness of these unintended mutations. Where relevant in our work, we will check for these outcomes using the new computational pipeline we have developed«, says Alanis-Lobato.
As technologies to edit and then analyse edited genomes advance, it is likely that more surprising outcomes are in store, but Alanis-Lobato reminds us that this need not be a bad thing:
»CRISPR-Cas9 is still a relatively new research tool. Like with many techniques, the more we use them, the more we learn about where they can be useful and where they have gaps. What is important is that researchers are aware of these gaps, so it may be possible to improve this technique and reduce the risks of both on and off target mutations.«
The open-source computational pipeline developed during this study can be accessed here.
Link to original article in PNAS:
Frequent loss-of-heterozygosity in CRISPR-Cas9–edited early human embryos.
Tags
ArticleInterviewNewsIn vivoRibonucleoprotein (RNP)Off-targetCRISPR-CasCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







