DNA Origami Unfolds as a New CRISPR Delivery System

The Japanese art of origami has stunned people for centuries by transforming a flat piece of paper into beautiful sculptures of intricate designs. In 2006, Paul Rothmund at Caltech took the art one step further and substituted the paper with DNA, and ever since, the genetic material has been used to create amazing 2D and 3D shapes. Now, a team led by Nobel Laureate Jennifer Doudna demonstrates how DNA origami can be used to fold an intact human gene into a package bound to CRISPR-Cas9 and guide RNAs that can be delivered together into the nucleus of human cells - ready for gene editing.
»One of the main questions that remains open in the CRISPR field is how we can introduce large pieces of DNA into specific places to, for example, create specific CAR T immunotherapies. Unfortunately, new technologies like prime editing cannot solve this problem since they're limited to inserting smaller pieces of DNA. So, we wanted to improve the efficiency of large insertions at targeted sites using CRISPR-Cas9,« explains Enrique Lin-Shiao, PhD, who is co-first author of an article that was published last month in Nucleic Acids Research.
He is a physicist and recently finished a postdoctoral fellowship in Jennifer Doudna's lab at the University of California, Berkeley. The paper's other co-first author, Wolfgang Pfeifer, PhD, is a nanoscientist at The Ohio State University, focusing on DNA origami structures. He explains:
»DNA origami structures cannot be found in nature, so we design them in a lab using software systems. We start with a piece of single-stranded DNA and then add so-called staple strands, i.e., 30-50 bases-long oligonucleotides. One staple strand has a complementary sequence to a part of the long DNA strand, but it also has complementary sequences to other parts of it. So, upon hybridisation, these parts will be brought into proximity. We don't do this only one time but add, let's say, hundreds of staple strands to create whatever shape we want.«
DNA nanostructures bring terminal ends together
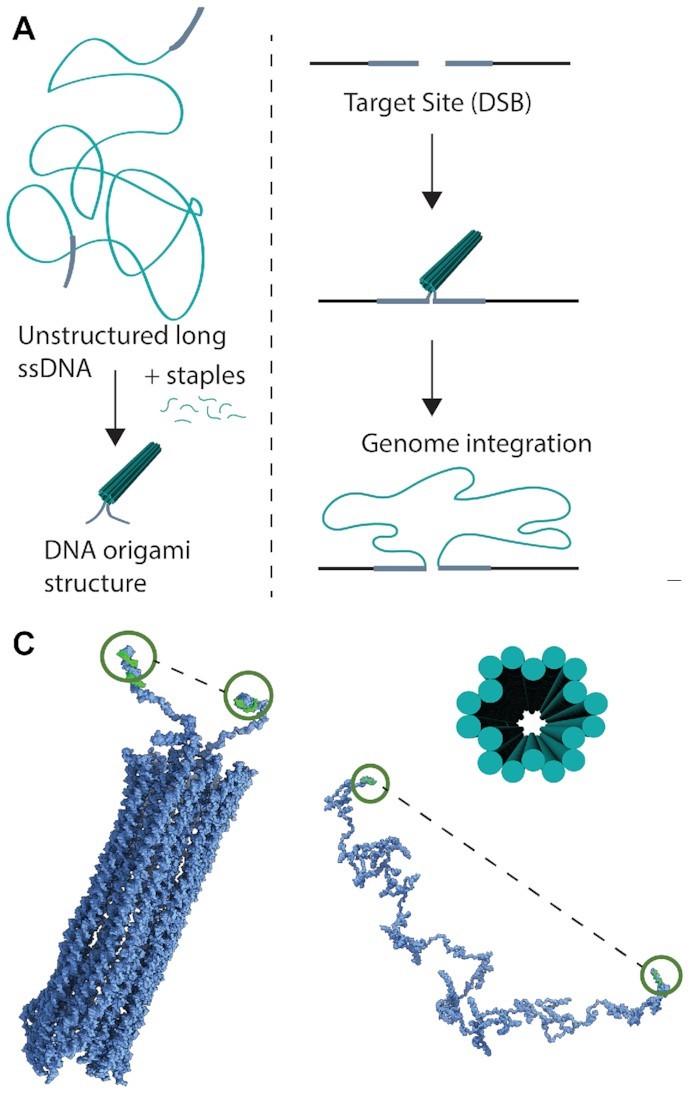
In the study, the two researchers and their colleagues used DNA encoding genes to create different origami nanostructures. Then they tested the ability of these structures to function as homology-directed repair (HDR) donor templates for precise, large genomic insertions using CRISPR–Cas9. First off, they used a 2716-nucleotide single-stranded DNA (ssDNA) scaffold encoding mNeonGreen. Each terminal of the DNA strand contained 100-nucleotide homology arms that matched the sequences flanking the intended CRISPR–Cas9 cleavage site (Figure 1).
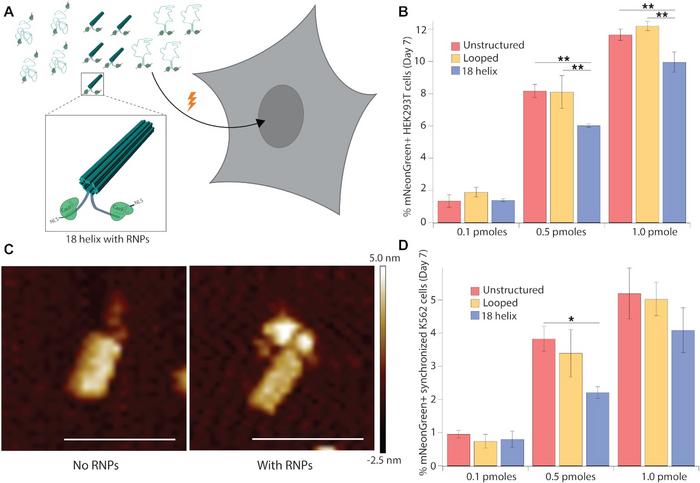
Initially, the unstructured ssDNA strand was compared to two other structures. One was a simple, looped structure where only the terminal homology arms were brought together by staple strands. The other was a compact DNA origami structure with 18 helices bundled into a hollow tube. Atomic force microscopy and molecular dynamics simulations revealed that structuring decreased the distance between the terminal ends to 29 nm, down from 109 nm in the unstructured ssDNA.
The different DNA structures were then delivered to HEK293T cells. Either of two delivery methods were used: transfection with plasmids encoding CRISPR–Cas9 and sgRNAs, or electroporation with ribonucleoproteins (RNPs). In both cases, the sgRNA targeted an intergenic site on human chromosome 9. After seven days, genome integration efficiency was analysed by flow cytometry for mNeonGreen-expressing cells. The results showed that both delivery methods allowed the 18-helix DNA nanostructures to enter the nucleus and become integrated into the genome. Integration of 18-helix nanostructures, however, occurred at lower frequencies relative to unstructured and looped DNA.
»We were a little bit disappointed that this first design wasn't immediately leading to the best integration. Many parameters must be optimised to get to that goal, and many follow-up experiments are needed to create designs that can lead to better integration and knock-in efficiency. For example, we want to know if packaging the gene into a specific shape can protect it from partial integration, which is sometimes a problem with single-stranded DNA. We also want to know if the integration efficiency of structured DNA is lower because it cannot open in time for HDR to happen,« Enrique Lin-Shiao says.
Packaged DNA is shuttled into the nucleus
To improve integration, the researchers added truncated Cas9 target sequences to the terminal arms of template DNA. This allowed RNPs to bind to the structures, so the nuclear localisation signals of Cas9 could facilitate transport into the nucleus. Atomic force microscopy imaging showed that CRISPR–Cas9 RNPs indeed bound to the 18-helix nanostructures. By also doubling the amount of each template from 0.5 pmol to 1.0 pmol, the proportion of mNeonGreen-expressing HEK293T cells after electroporation increased from around 4% without bound RNPs to 10% with bound RNPs (Figure 2).
Next, the team wanted to see if DNA nanostructures could be used as an approach to correct any mutation in a gene by inserting the entire wild-type DNA sequence. This might be useful for diseases like Duchenne muscular dystrophy (DMD) that can arise from several mutations or deletions, so you otherwise would have to create a specific therapy for each mutation.
“Large genes like dystrophin or factor VIII could be exciting targets for treating DMD and haemophilia, respectively, using our approach”Enrique Lin-Shiao
The approach was tested using the interleukin 2 receptor alpha (IL2RA) gene, where several mutations can lead to immunodeficiencies. First, a 3.5-kb multigene cassette containing exons 2-8 of IL2RA and reporter genes was packaged into nanostructures with terminal shuttle sequences. The nanostructures and RNPs that were targeted for insertion into exon 1 of the native gene were then electroporated into primary human T cells. Even with this large construct, the 18-helix DNA nanostructure facilitated knock-in in around 15% of cells, which was comparable to unstructured and looped DNA. Indeed, Enrique Lin-Shiao believes that the approach could work for genes of any size.
»I don't think there's a size limitation, and people working in the DNA origami field have typically been working with pieces of DNA that are about 8 kb long. But, it is also possible that even if you don't see a big difference for 3 kb, because the efficiency is already relatively high, there might be much more benefits to folding the DNA into nanostructures if you go much larger. So, large genes like dystrophin or factor VIII could be exciting targets for treating DMD and haemophilia, respectively, using our approach,« he says.
DNA origami and virus-like particles are a perfect match
Electroporation is useful for CRISPR experiments or therapies ex vivo, but the delivery method is not suitable for in vivo therapies. Accordingly, the team tested the efficiency of mNeonGreen integration using delivery with virus-like particles (VLPs) that are also applicable in an in vivo setting. These experiments showed that integrating 18-helix nanostructures was significantly less effective using VLPs (5% expressing cells) than electroporation (13% expressing cells). However, 18-helix nanostructures faired almost three times better than unstructured or looped DNA when delivered by VLPs. In contrast, all three DNA structures were equally efficient when delivered by electroporation (Figure 3).
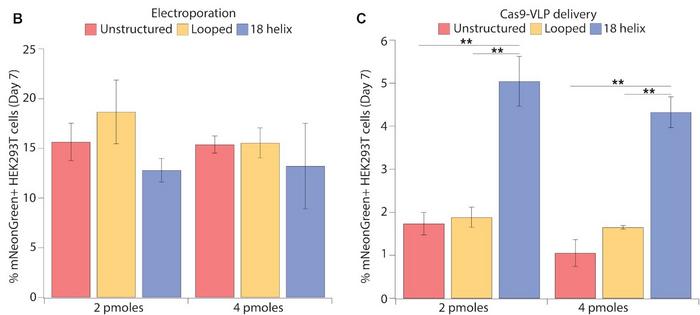
»We are not entirely sure why the 18-helix nanostructure is working so much better than unstructured DNA with VLPs. We don't know if it's because the folding allows more of the DNA to go into the VLPs, or if the VLPs are carrying a lot of nanostructures on their surface. These are open questions,« says Enrique Lin-Shiao.
The most significant potential for DNA origami in the CRISPR field might be the possibility of creating therapies for genetic diseases like DMD, where there are many disease-causing mutations and deletions. At present, there is no single strategy that can treat all DMD patients, but this new technology might create a more general approach to benefit all those patients at once. But there are also many other exciting possibilities, according to Wolfgang Pfeifer:
»Based on things we learned in this study, this opens up for a whole field of potential applications for DNA origami like, for example, biosensors that can target specific loci in the cytoplasm or nucleus. However, there is so much we can still learn, and it is so close, within reach.«
Link to the original article in Nucleic Acids Research:
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsin vivoIn vivoNon-viralElectroporationLipid-based nanoparticleRibonucleoprotein (RNP)Duchenne Muscular Dystrophy, DMDHaemophiliaCas9
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University