Engineered deaminases enable precise RNA base editing
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Base editing has emerged as a powerful approach for modifying genetic material without the double-strand breaks typically induced by conventional CRISPR-Cas nucleases. While DNA base editing has advanced considerably, RNA base editing offers distinct advantages – the modifications are transient rather than permanent, providing a potentially safer avenue for therapeutic interventions.
“Strategically designed point mutations in the NTD and CTD of APOBEC3A can significantly decrease off-target effects while maintaining sufficient editing efficiency”Han et al.
The field has seen progress with A-to-I editing using ADAR enzymes, but achieving efficient C-to-U transitions has remained challenging. Previous attempts to harness APOBEC enzymes for CRISPR-Cas13-based systems, such as CURE, have achieved only limited success, with editing efficiencies of around 20% in vivo. The fundamental problem stems from APOBECs' preference for single-stranded RNA, which conflicts with the double-stranded structures formed between target mRNA and guide RNAs in CRISPR-Cas systems.
To overcome these limitations, the researchers took a three-pronged approach. First, they optimised the Pumilio and FBF (PUF) protein domain – a programmable RNA-binding protein where each repeat recognises a specific nucleotide – by introducing a short leucine-proline peptide, which improved structural stability and increased editing efficiency from 69.7% to 82.3%. Second, they introduced strategic point mutations in APOBEC3A to reduce dimerisation and RNA binding affinity, which decreased off-target effects whilst maintaining on-target activity.
»Strategically designed point mutations in the NTD and CTD of APOBEC3A can significantly decrease off-target effects while maintaining sufficient editing efficiency,« the authors note in their Nature Communications paper.
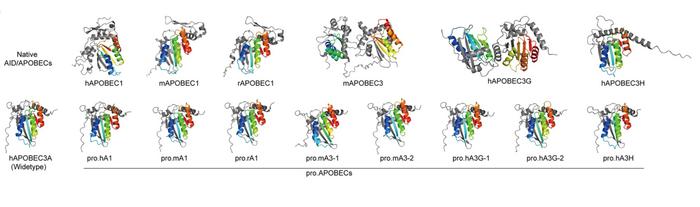
The third and most innovative step involved AI-assisted protein engineering to create Professional APOBECs, or ProAPOBECs. Using AlphaFold2 to analyse structures across the AID/APOBEC family, they recognised that whilst the catalytic deaminase domain is conserved, the terminal domains vary considerably. The researchers systematically combined deaminase domains from human and rodent APOBEC1 and APOBEC3 proteins with the terminal domains of human APOBEC3A, generating a library of chimeric enzymes with altered sequence preferences (see Figure 1).
When incorporated into the REWIRE system (RNA editing with individual RNA-binding enzyme), which uses PUF proteins rather than CRISPR-Cas components for target recognition, these ProAPOBECs demonstrated unprecedented versatility. Different variants showed strong C-to-U editing activity in GC, CC, AC and UC sequence contexts – a substantial expansion beyond the UC-only specificity of natural APOBEC3A. CU-REWIRE5.1 (CU5.1) achieved efficient editing in GC contexts, while CU5.15 showed activity in CC contexts, and CU5.3 and CU5.17 could edit cytosines in essentially any sequence context.
The researchers validated their system with several disease-relevant targets in vitro, successfully editing the c.C526T variant in APOE with approximately 90% efficiency. In this target, existing editors showed no activity. They also demonstrated editing of mutations in SOD1 associated with amyotrophic lateral sclerosis and in rhodopsin causing retinitis pigmentosa, achieving efficiencies around 85%.
The translation to in vivo applications proved the technology's therapeutic potential. The team packaged CU5.21 into AAV8 vectors targeting Pcsk9 in the mouse liver, introducing a premature stop codon at position C832. Four weeks after injection, treated mice showed over 70% reduction in Pcsk9 mRNA levels, accompanied by decreases in PCSK9 protein and serum cholesterol levels.
Transcriptome-wide RNA sequencing identified approximately 400 C-to-U editing events. Notaby, deep sequencing of genomic DNA revealed no detectable C-to-T mutations even at sites with 96% RNA editing efficiency.
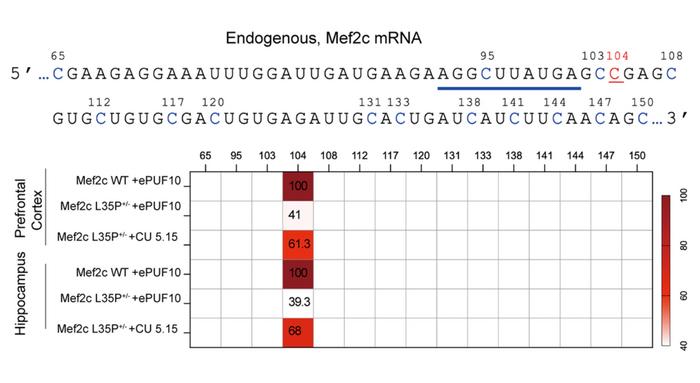
The second in vivo application targeted the brain using an autism spectrum disorder mouse model carrying the Mef2c L35P mutation (c.T104C), which causes severe autism in humans. The researchers packaged CU-REWIRE5.15 into blood-brain barrier-crossing AAV-PHP.eB vectors.
Four weeks after injection, RNA sequencing revealed C-to-U editing rates of 61.3% and 68% in the prefrontal cortex and hippocampus, respectively, with no bystander editing (see Figure 2). MEF2C protein levels were fully restored, and treated mice showed normalised social behaviours across multiple assays. Remarkably, the lifespan of treated mice approached that of wild-type animals – an improvement not observed with previous DNA editing approaches in this model.
»RNA base editing facilitated by CU5.15 provides a potent tool for correcting genetic mutations in the brain and has potential therapeutic applications for ASD and other genetic disorders,« the authors emphasise.
The system's design offers several advantages over CRISPR-Cas-based approaches. The use of mammalian-derived proteins rather than bacterial Cas enzymes minimises potential immune responses in therapeutic settings. The PUF-based targeting provides flexibility without requiring PAM sequences or forming extensive RNA duplexes that might inhibit APOBEC activity. Immunofluorescence confirmed that CU-REWIRE constructs predominantly localise to the cytoplasm, reducing potential genomic interactions.
The work demonstrates how AI-assisted protein engineering can rationally expand the capability of enzymes. By analysing over 8,000 eukaryotic AID/APOBEC genes and using AlphaFold2 predictions to identify structurally compatible domain combinations, the team created thousands of potential ProAPOBEC variants with altered substrate specificities. The high editing rates achieved – up to 96% in vitro and 68% in vivo – approach levels necessary for phenotypic correction. The absence of detectable DNA mutations addresses a critical safety concern, whilst the relatively contained transcriptome-wide off-target profile appears manageable, particularly as these modifications are transient.
The study was conducted by Wenjian Han and Bo Yuan from Shanghai Jiao Tong University School of Medicine and Fudan University, respectively, and their colleagues. It was published in Nature Communications on 4 November 2025, and was led by Zilong Qiu and Zefeng Wang.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleCMN HighlightsNewsDelivery - BrainIn vivoAdeno-associated virus (AAV)Familial Hypercholesterolemia, FHCardiovascular diseasesAI in genome editingBase editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.