Explainer: What Are Base Editors and How Do They Work?
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
What is base editing and what is it used for?
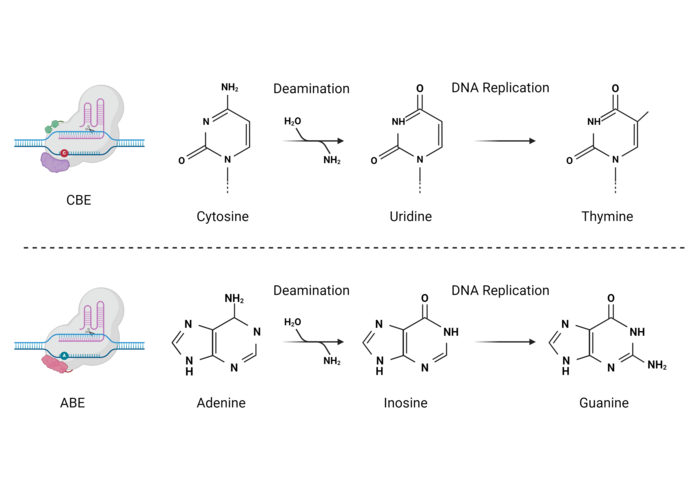
Base editing is a gene-editing technology described initially by the laboratories of Drs. David Liu and Akihiko Kondo in 2016 (1,2). Base editing combines the powerful DNA-scanning and sequence-identification capabilities of the CRISPR-Cas9 system with a deaminase enzyme, which introduces single nucleotide polymorphisms (SNPs) by chemically altering the target DNA sequence without the intentional generation of a DNA double-strand break (DSB). This chemical modification, known as deamination, consists of the removal of an amino group from a nucleotide, which after DNA repair or replication results in the installation of a new base (see Figure 1).
The first base editors developed in 2016, called cytosine base editors (CBEs), can substitute a cytosine base for a thymine (C→T), while newer base editors called adenine base editors (ABEs) are able to introduce adenine-to-guanine (A→G) substitutions (3). More recently, base editors capable of cytosine-to-guanine (CGBEs) or simultaneous adenine and cytosine (ACBEs) substitutions have also been developed but their application is still limited to a handful of studies (4,5).
Base editing is an attractive way to edit DNA, because it greatly reduces some of the major risks associated with conventional, DSB-mediated gene editing: the heterogeneity of insertions and deletions (INDELs) at on-target sites, the increased potential for DSB-related chromosomal aberrations, and cell toxicity.
What are the components of a base editor?
Base editors are generated by coupling two separate proteins capable of very specific functions, as illustrated in Figure 2. The components of a typical base editor include:
- Cas9 nickase (nCas9): This is a version of Cas9 that has been modified through mutations in one of the two main amino acid residues responsible for the DNA cleavage activity of Cas9. Thus, nCas9 is still able to pair with a gRNA and find the DNA sequence complementary to the gRNA spacer, but is only able to nick one strand of the DNA.
- Nucleoside deaminase: This is an enzyme capable of removing an amino group from a specific type of nucleoside. The deaminase fused to the nickase – be it a cytosine deaminase (e.g. APOBEC) or an adenosine deaminase (e.g. engineered TadA) – will dictate whether the base editor is a CBE or an ABE. CBEs contain cytosine deaminases, while ABEs typically contain adenine deaminases (though several groups have recently engineered CBEs from the conventional TadA adenosine deaminase domain (6-8)).
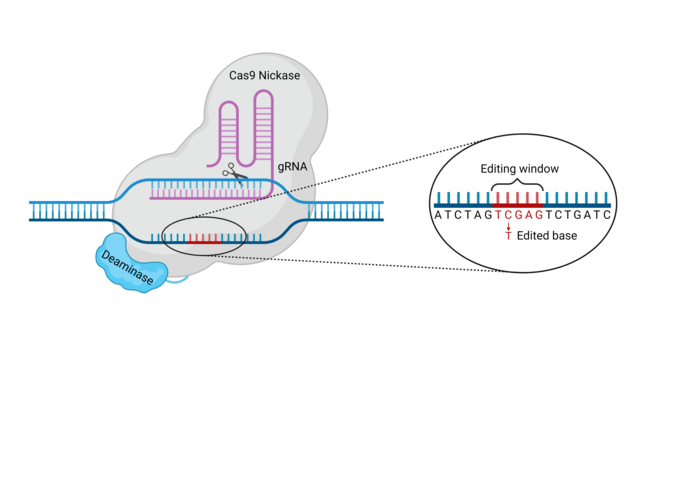
The Cas9-deaminase fusion protein complex is targeted to a specific DNA locus by a guide RNA (gRNA). Once the base editor has bound to its target sequence, the deaminase can modify bases within the exposed non-target strand (NTS) of the target site R-loop. The area of the target locus in which bases can be modified is called the base editing window (see Figure 2). Depending on the specific intended modification, other factors may be included in the base editor complex to increase efficiency or alter the outcome of the base substitution.
**CRISPRMED25 - The 2nd CRISPR Medicine Conference, Copenhagen, Denmark, April 8-11, 2025**
Learn about the latest discoveries in CRISPR Medicine at the CRISPRMED25 Conference in Copenhagen, Denmark, April 8-11, 2025.
How does base editing work?
Once the base-editing complex recognises the target DNA sequence complementary to the gRNA spacer, Cas9 mediates the denaturing of the double-stranded DNA (dsDNA) to form a structure known as an R-loop, which exposes a short stretch of single-stranded DNA (ssDNA) in the non-complementary strand to the activity of the deaminase (see Figure 3; also called the non-target strand). This allows the deaminase to chemically modify bases inside the base editing window, which is normally 5-10bp long depending on the deaminase, and somewhat distal from the protospacer adjacent motif (PAM) within the target site.
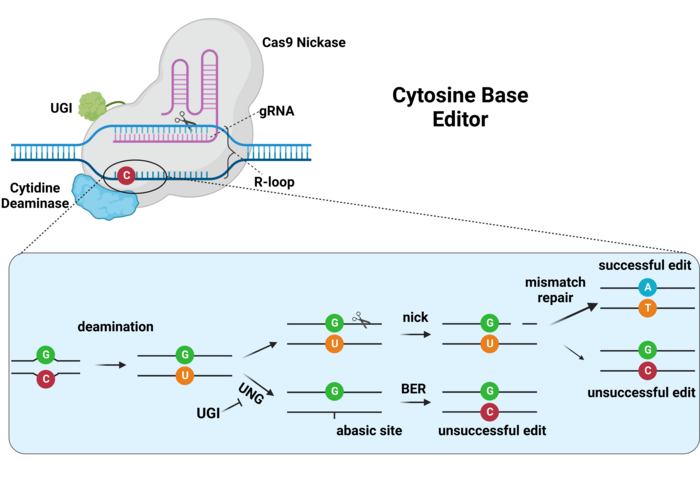
The first generation of base editors encountered severe efficiency issues that had to be solved. For example, CBEs mediate the deamination of cytosines, generating a uracil (U) base, which can then be read by the cell’s DNA polymerases as thymine. However, human cells are very efficient at DNA repair, and especially at dealing with deaminated bases via the base-excision repair (BER) pathway. Thus, uracil can be recognised and eliminated by the enzyme uracil DNA N-glycosylase (UNG) during the initiation of BER. This drastically limits the efficiency of C-to-T edits and can lead to reduced C-to-T purity (and increased C-to-A or C-to-G edits). To overcome this issue, second-generation CBEs included a third component, namely a uracil glycosylase inhibitor (UGI) fused together with Cas9 and the cytosine deaminase. UGI inhibits the action of UNG on the edited DNA, thus conserving the uracil base and achieving 3-fold higher editing efficiency and purity (Ref. 1, and see Figure 3).
In addition to the above, the DNA mismatch repair mechanisms of the cell are tasked with detecting and resolving any incorrect nucleotide pairings into correct pairings (A-T and C-G). Since the deamination of a cytosine, which will be paired with a guanine, will lead to an incorrect T-G pairing, this will be repaired by the cell, either by substituting the U for a C (C-G pairing) or by substituting the G for an A (T-A pairing). Considering that the cell does not know which of the two bases is correct, the probabilities of each repair outcome are equal. To increase editing efficiency, the catalytically-dead version of Cas9 (dCas9) that was used in the first-generation CBEs was substituted with the Cas9 nickase (nCas9) version that we find nowadays in nearly all base editors, which is only able to nick the complementary strand of the DNA. This cut in the complementary (=unedited) strand, directs the mismatch repair machinery to utilise the base-edited strand as a template for repair, thus biasing the repair towards the introduced edit, and further improving efficiency (Ref. 1, and see Figure 3).
CRISPRMED26, April 14-16th, 2025 - Stay tuned...

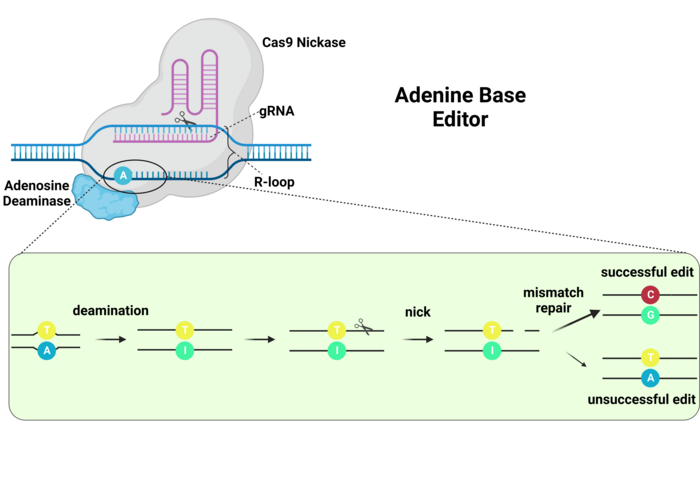
ABEs work in a very similar way. In this case, the deamination of adenine generates an inosine residue, which is interpreted by the DNA polymerases as a guanosine. Thus, an A-to-G edit is achieved (Ref. 3, and see Figure 4). However, no UGI or equivalent inhibitor is necessary since inosine residues are not excised like uracil residues.
One step that was very important for the generation of ABEs was making an adenine deaminase that could edit the DNA, since naturally occurring adenine deaminases only edit RNA. Directed mutagenesis efforts were successful in generating a version of the E.coli tRNA adenosine deaminase (TadA) that can edit the DNA. Interestingly, editing efficiency was higher when the ABEs included one mutated TadA (TadA*) and one wildtype enzyme (3). ABEs are very important because G-C to A-T mutations represent the most frequently reported pathogenic point mutations, thus this type of base editing has the greatest potential for clinical applications. More recent ABEs have been developed that have improved editing efficiencies (9,10).
How has base editing been used so far?
Several fascinating therapeutic approaches using base editing have been developed all over the world in the last few years. For example, in August 2022, Verve Therapeutics initiated a clinical trial using an intravenously delivered lipid nanoparticle that carries an ABE mRNA and a gRNA programmed to disrupt and silence the PCSK9 gene in the liver of patients with familial hypercholesterolemia (11).
More recently, two independent research groups have shown the potential of base editing to treat spinal muscular atrophy in patient-derived cells and mice by introducing an A-to-G edit in the SMN2 gene to increase its expression and compensate for the lack of expression in the mutated paralogous SMN1 gene (12,13).
Another interesting use of base editing has been to simultaneously disrupt the CCR5 and CCRX4 receptors of CD4+ T cells. This has been shown to be an effective method of rendering those cells immune to HIV infection and has the potential to become an effective way to prevent HIV infection/reinfection (14).
Although major advances have been made within base-editing technology so far, we have likely only scratched the surface of its potential in research and therapeutics.
What are the limitations of base editing?
Despite its advantages over other gene-editing technologies, base editing presents some limitations that are currently being addressed by scientists all over the world.
Some of the limitations are similar to those of conventional Cas9-based editing approaches regarding targeting, off-target editing and delivery. Thus, already existing Cas9 variants that were developed to solve those issues can be used to construct base editors able to overcome the same issues. When it comes to in vivo delivery, the increased size of the base editor complex compared to regular Cas9 complicates its delivery. To address this hurdle, the intein system, which is used to package other Cas9 products into adeno-associated viral vectors, has also been used for in vivo delivery of base editors. However, other, base editing-specific limitations have arisen:
- Requirement for precise positioning of the base editor edit window: To achieve efficient editing, base editors must be capable of recognising PAMs at an appropriate distance upstream of the intended edit (to position the edit window accordingly). Fortunately, various Cas9 PAM variants have been developed that utilise alternate PAMs, enabling more flexible base editing across the genome (15-17).
- Unwanted edits in the editing window: In some cases, when trying to edit a specific base, for example a cytosine, another cytosine inside the editing window can be deaminated as well, leading to unwanted effects. To avoid this, base editor variants with narrower editing windows have been developed by introducing mutations in the deaminase enzymes.
- Indel generation: Since new-generation base editors introduce single-strand nicks in the non-edited strand, in some cases the excision of an edited base by the BER can lead to a DSB. The repair of DSBs leads to the generation of INDELs. Strongly inhibiting the BER pathway is one approach to avoid DSB formation.
- Off-target RNA editing: An interesting and unexpected observation made by some scientists was that base editors could also produce unwanted edits to RNA, independently of Cas9 activity, which could lead to unpredictable effects. Efforts to solve this issue have focused on engineering the deaminase protein to reduce its activity on the RNA.
Stay tuned for future explainers about various gene-editing technologies. In the meantime, you can find all of our coverage on base editors right here.
Cited literature:
- Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016 May 19;533(7603):420-4. doi: 10.1038/nature17946.
- Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016 Sep 16;353(6305):aaf8729.
- Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017 Nov 23;551(7681):464-471.
- Kurt IC, Zhou R, Iyer S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021 Jan;39(1):41-46.
- Huang TP, Newby GA, Liu DR. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc. 2021 Feb;16(2):1089-1128.
- Neugebauer ME, Hsu A, Arbab M, et al. Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat Biotechnol. 2023 May;41(5):673-685.
- Lam DK, Feliciano PR, Arif A, et al. Improved cytosine base editors generated from TadA variants. Nat Biotechnol. 2023 May;41(5):686-697.
- Chen L, Zhu B, Ru G, et al. Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat Biotechnol. 2023 May;41(5):663-672.
- Richter MF, Zhao KT, Eton E, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020 Jul;38(7):883-891.
- Gaudelli NM, Lam DK, Rees HA, et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol. 2020 Jul;38(7):892-900.
- Musunuru K, Chadwick AC, Mizoguchi T, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021 May;593(7859):429-434.
- Alves CRR, Ha LL, Yaworski R, et al. Base editing as a genetic treatment for spinal muscular atrophy. bioRxiv [Preprint]. 2023 Jan 21:2023.01.20.524978.
- Arbab M, Matuszek Z, Kray KM, et al. Base editing rescue of spinal muscular atrophy in cells and in mice. Science. 2023 Apr 21;380(6642):eadg6518.
- Knipping F, Newby GA, Eide CR, et al. Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol Ther. 2022 Jan 5;30(1):130-144.
- Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020 Apr 17;368(6488):290-296.
- Nishimasu H, Shi X, Ishiguro S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018 Sep 21;361(6408):1259-1262.
- Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015 Jul 23;523(7561):481-5.
Other relevant literature:
- Huang TP, Newby GA, Liu DR. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc. 2021 Feb;16(2):1089-1128. doi: 10.1038/s41596-020-00450-9.
- Porto EM, Komor AC. In the business of base editors: Evolution from bench to bedside. PLoS Biol. 2023 Apr 12;21(4):e3002071.
- Jeong YK, Song B, Bae S. Current Status and Challenges of DNA Base Editing Tools. Mol Ther. 2020 Sep 2;28(9):1938-1952.
Manel Llado is a gene editing expert and science writer/communicator based in Vienna, Austria
We acknowledge Dr. Benjamin P. Kleinstiver (Mass General Hospital and Harvard Medical School) for advice and input.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







