First CRISPR Manipulation of a Mammalian Microbiome via a Phage-Bacteria Pair
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The Human Microbiome Project, which began in 2007, shed light on the makeup of the human microbiome during health and disease. The past five years in particular have seen an explosion in gut microbiome research, including the application of genetic tools including CRISPR to interrogate the microbiome to increase our understanding of the mechanistics behind microbe-microbe and microbe-host interactions, and to potentially reshape it for therapeutic purposes.
Given their natural propensity for bacterial infection, engineered bacteriophages are seen as prime delivery vehicles for CRISPR-Cas reagents to target bacterial pathogens. To date, bacteriophages have been applied as antimicrobials to target Staphylococcus aureus, Clostridium difficile or Escherichia coli in mouse skin or gut infection models, with success. However, until recently the application of M13 phage (see fact box) to deliver genetic constructs to bacteria within the gut microbiome had not been attempted.
In a recent study published in Cell Reports, Dr Peter Turnbaugh’s research group at the University of California, San Francisco, describe a system that exploits conjugation between the M13 phage and E. coli to deliver a CRISPR-Cas9 construct to specific bacteria within the mouse gut.
This work provides a precise targeting mechanism that can deliver CRISPR reagents to a gut pathogen, and addresses a major deficit in the field, as Dr Turnbaugh explains: »One of the major limitations that we have in the microbiome field is that we lack the equivalent of a knockout mouse. When we study these complex microbial communities, it is hard to know which members of the community are necessary for any given function. We turned to the M13 bacteriophage and E. coli pair as a starting point to probe whether it was feasible to use M13 to deliver CRISPR reagents in the gut.«
Bacterial Conjugation and M13 Phage
E. coli cells carrying the F plasmid, which encodes a Type IV secretion system, form thin flexible filaments called F pili that initiate contact with a recipient cell. This establishes a mating pair and supports the transfer of genetic material from the E. coli (donor) to the recipient cell. The M13 phage exploits this mechanism: it targets the tip of the pili, effectively conjugating with E. coli instead of a recipient cell. This leads to the formation of a phage-bacteria mating pair, which uses the conjugative contact to infect a target bacterial cell.
Leveraging a bacteriophage to study microbial community function
A previous study used an engineered M13 bacteriophage carrying CRISPR-Cas reagents to target bacterial infection in an insect model. Additional studies, which did not include CRISPR in their approach, used the M13 bacteriophage to target the gut pathogens E. coli and Helicobacter pylori in mouse infection models. While these approaches employed the M13 bacteriophage as a delivery system to fight bacterial infections, the new platform developed by Dr Turnbaugh’s group represents the first application combining the M13 bacteriophage and CRISPR for use in the mouse gut.
According to Dr Turnbaugh, the system can be used to clarify which members of a complex natural community are responsible for a given phenotype by precisely removing species or genes from a microbial community:
»This work has set up the tool, and we want to use it to gain a greater understanding of the microbiome, either by affecting something that E. coli is doing already or by forcing it to do something novel.«
Phage-mediated DNA delivery in the mouse gut
To first assess the ability of the M13 bacteriophage to successfully deliver a payload to E. coli, the team tested the delivery of a plasmid containing a β-lactamase gene, which confers resistance to β-lactam antibiotics such as ampicillin or carbenicillin. To encapsulate a desired plasmid into the M13 phage, E. coli cells are transformed with the plasmid and cultured. The M13 phage is then added to the culture, where it infects the bacteria and picks up the plasmid. The phage is subsequently collected from the culture supernatant.
Mice used in this study were initially treated with streptomycin – an antibiotic commonly used in such experiments to eradicate the native microbiota and to allow engraftment of the strain under study - to ensure successful colonisation with E. coli. The mice were subsequently dosed orally with the M13 phage harbouring the β-lactamase gene and treated with carbenicillin.
Successful conjugation could then be assessed by the presence of carbenicillin-resistant faecal E. coli isolates after antibiotic treatment. Since E.coli is susceptible to β-lactam antibiotics, the resistance marker makes it easy to identify E. coli cells successfully infected with the phage, as Dr Turnbaugh explains: »We started with a simple delivery of displaying a plasmid that has an antibiotic resistance gene. When we dosed the mice with that antibiotic, we could show that the E. coli had been targeted by the phage.«
The next step was to determine if the system could target a particular bacterial strain using a CRISPR-Cas9 construct. To test this, the team engineered isogenic E. coli strains expressing either the GFP (green fluorescence) or the mCherry (red fluorescence) fluorophore and used those genes as deletion targets for their M13-based CRISPR system. This setup allowed them to assess the efficiency and target specificity of the system easily in vitro by checking for the loss of fluorescence. Whole-genome sequencing of the affected clones confirmed chromosomal deletions encompassing the targeted gene (Figure 1).
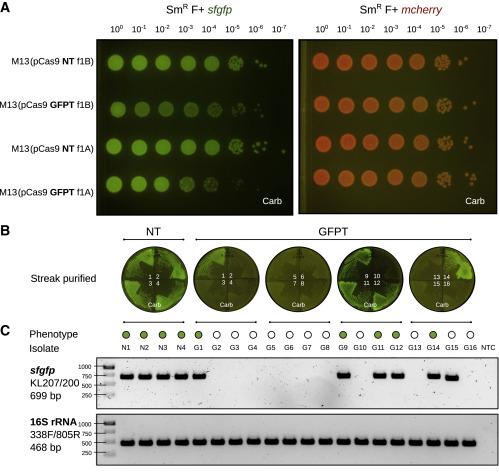
The group then went on to assess whether they could accomplish a sequence-specific depletion of E. coli within the mouse gut microbiome. Mice that were colonised with an E. coli strain carrying genes for both GFP and mCherry were dosed with M13 carrying a GFP-targeting CRISPR-Cas9 plasmid. Over the course of 2 weeks, the team observed loss of the GFP signal and maintenance of the mCherry signal, findings that were validated by flow cytometry, microbiological streaking and sequencing of isolated E. coli clones. These findings indicated successful CRISPR-Cas9 delivery and specific GFP-encoding gene targeting that allowed strain-specific depletion (Figure 2).
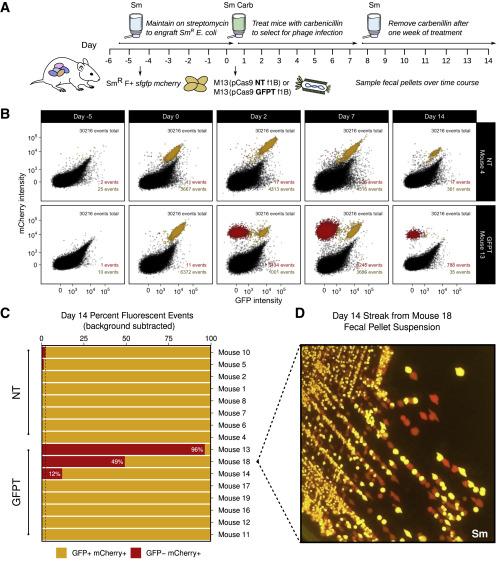
Of note, the CRISPR-Cas targeting system designed by Turnbaugh’s team mimics a natural configuration found in bacterial genomes. This means that targeting is carried out by a spacer flanked by repeat sequences rather than a guide RNA.
The team also assessed phage viability throughout the gastrointestinal tract, since bacteriophage loss is a common challenge in phage-based gut delivery systems. Although Dr Turnbaugh’s team found the M13 phage to be significantly tolerant to acid pH, they still detected phage loss in the mouse model, with 1 x 106 viable plague-forming units remaining from an input of 6 x 1013 plague-forming units. This observation suggests that other digestive components such as secreted enzymes or bile acids may be impacting phage survival in the gut, and finding ways to counteract this issue should increase the viability of phage-mediate therapies in general.
Is targeting pathogen virulence in the gut the only strategy?
The proof-of-principle platform developed by Dr Turnbaugh’s group means that it is now possible to evaluate the success of genome editing in the mouse gut using a straightforward fluorophore system. The setup can be easily adapted to assess the editing efficiency of other plasmid-borne CRISPR reagents or even applied to other phage-bacteria pairs.
When thinking of CRISPR as an antimicrobial, the optimal outcome would be to wipe out the whole pathogenic population. However, according to Dr Turnbaugh, this work shows that gene depletion may not always lead to cell death: »This study and others before have shown that it is not that clean, some bacteria are able to repair the double-strand break by homologous recombination.«
Additionally, selective pressure by an antimicrobial CRISPR system can cause resistance to emerge in the targeted population. »In both the in vitro experiments and in mice, we saw escape from the system through various mechanisms«, Dr Turnbaugh says. These included target site mutations, loss of the CRISPR array spacer or even loss of the entire CRISPR-Cas9 system.
So instead of wiping out entire pathogen populations, Dr Turnbaugh envisions a strategy where CRISPR is used to promote positive microbiota effects, as he explains: »It might be useful to target some other members of the community with the CRISPR system and to use the antibiotic to kill the pathogen«, arguing that »Antibiotics are already being used and for the most part they have good safety profiles. If you could give a pulse of antibiotics, together with a stable CRISPR edit, this could be a very effective strategy.«
To Dr Turnbaugh, CRISPR technology also holds promise as a metabolic reprogramming tool that may be used to improve human health in ways besides treating infections: »We really want to target other genes of interest besides genes present in pathogens. In particular, we are interested in microbial metabolism, including enzymes that E. coli expresses that can help with the effects of drugs or components of our diet.«
Translating animal model data to human use cases
Going forward, Turnbaugh’s team will work on improving the efficiency of the system and devise ways to decrease the escape of targeted cells. Eliminating the use of an antibiotic to monitor for DNA delivery to E. coli is also a future objective.
The challenge of finding other phage-bacteria pairs is among Dr Turnbaugh’s future plans: »We need an M13 equivalent for all the other [bacterial] species we are interested in. Having that would take us somewhere really exciting, but to do so, it is necessary to identify phages that target other bacteria of interest. Only very few of the species in the microbiome have been studied so far, in terms of their genetics or metabolism,« Dr Turnbaugh says.
Translating microbiome knowledge from mice to the human host is an important challenge that needs to be addressed. One issue is that mice are typically treated with antibiotics that decrease microbial community complexity. Dr Turnbaugh has a plan to solve this: »It would be great to start with human microbial communities that we could either target in vitro or using a model, where we take a complex community from a human and use it to colonise germ-free mice. That way it is possible to maintain a much more complex, human-derived microbial community in mice.«
This work is a good illustration of the importance of a good basic science foundation to answer complex and dynamic research questions, and the findings should open new doors in microbiome research. Dr Turnbaugh concludes: »A lot of excitement comes from the pairing of phage and CRISPR systems because it provides a much more precise way to direct CRISPR reagents. And you can incorporate the platform into traditional phage therapy setups, you just have to choose the strain to target.«
Link to the original article in Cell Reports:
Pedro Pais is a molecular biologist and microbiologist based in Portugal.
Tags
ArticleInterviewNewsIn vivoInfectious diseaseBacterial InfectionMicrobiomeCRISPR-CasCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







