First In Vivo Prime Editing Study Corrects Pathogenic Mutations in Mice
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Prime editing (PE) has been under the spotlight recently as a safer and more precise alternative to traditional CRISPR-Cas9 gene editing. Using a Cas9 nickase and a reverse transcriptase, prime editors can create small insertions and deletions without creating double-strand breaks (DSBs) in DNA, and without requiring a donor template for repair.
Since prime-editing systems were first described by Dr. David Liu’s lab at Harvard in 2019, gene therapy researchers have been eager to see the technique used in animal models.
In a study published recently in Nature Biomedical Engineering, researchers from Hyongbum Henry Kim’s lab at the Republic of Korea’s Yonsei University College of Medicine used PE to correct mutations responsible for two different genetic diseases in adult mice, with no evidence of off-target effects. The paper is the first to demonstrate the use of prime editing in vivo, and provides a compelling case for the superiority of PE over other gene-editing methods.
Dr. Dong Hyun Jo, formerly a postdoctoral fellow in Professor Jeong Hun Kim’s lab, now an assistant professor at the Seoul National University College of Medicine, has spent several years attempting to use gene-editing technologies to treat paediatric retinal diseases caused by pathogenic mutations.
Tackling Leber’s congenital amaurosis with the PE2 system
Jo’s current research is primarily focused on treating Leber’s congenital amaurosis (LCA). LCA is well known among the gene-editing community, as it was the first disease to be treated in vivo in human subjects using CRISPR-Cas9 gene-editing technology in the ongoing BRILLIANCE clinical trial.
Jo and his colleagues in the Kim lab originally tested the therapeutic potential of SpCas9 in a mouse model of LCA, followed by similar attempts to treat the condition using an adenine base editing-mediated technique named CRISPR-PASS.
»Before gene therapy and genome editing, these diseases were thought to be ‘untreatable.’ I have been fascinated by the first demonstration of the therapeutic potential of Cas9 and base editing in an animal model of LCA. Both techniques have great potential but have specific limitations such as low efficiency (original Cas9-mediated HDR) and limited types of potential conversion (base editing). Just after the publication of the Liu lab’s prime editing paper, we decided to investigate the therapeutic potential of prime editing in vivo in animal models,« Jo explains.
There are now four different types of prime-editing systems. PE1 and PE2 use a Cas9 nickase fused to a reverse transcriptase and a prime-editing guide RNA (pegRNA). PE3 and PE3b are derivatives which use the PE2 system in conjunction with an additional single guide RNA (sgRNA). Interestingly, the team used not only one, but two prime-editing systems in the study, treating two different diseases.
To determine whether PE2 systems can be used to treat LCA, the team used the common rd12 mouse model of LCA. Editing components were injected directly into the retina and retinal pigment epithelium (RPE) of the mice, a treatment which corrected the mutation and resulted in significantly improved visual function.
Correcting hereditary tyrosinemia using PE3
The other genetic disease the authors tackle in the current study is hereditary tyrosinemia, an autosomal recessive condition caused by a loss-of-function mutation in the fumarylacetoacetate hydrolase (Fah) gene. The pathogenic mutation prevents patients from producing the fumarylacetoacetate hydrolase enzyme, which contributes to normal breakdown of the amino acid tyrosine. The resulting toxic buildup of tyrosine in the bloodstream leads to a range of health complications including liver failure.
»Most genetic diseases are untreatable with conventional treatment options, but impose a significant burden on children and their families. It is inevitable to investigate therapeutic options for these genetic diseases as clinician-scientists,« Jo remarks.
Along with attempting to provide novel treatment options for these conditions, one of the key aims of the study was to compare the results of prime editing with those achieved previously, both in Jo’s own lab and from other studies.
»We and other groups have investigated the effects of homology-directed repair, microhomology-mediated end joining, and base editing in both animal models [of hereditary tyrosinemia and LCA]. We speculated that we could compare the editing efficiency of prime editing with previous reports,« Jo adds.
The team used an FAHmut-mutmouse model of hereditary tyrosinemia type 1, delivering PE3 components via hydrodynamic injection. All the PE3-treated mice displayed amelioration of disease phenotype and survived until the end of the study period, while the control group showed weight loss and limited survival. The treated mice also displayed correction of the FAH mutation in a fraction of hepatocytes (Figure 1).
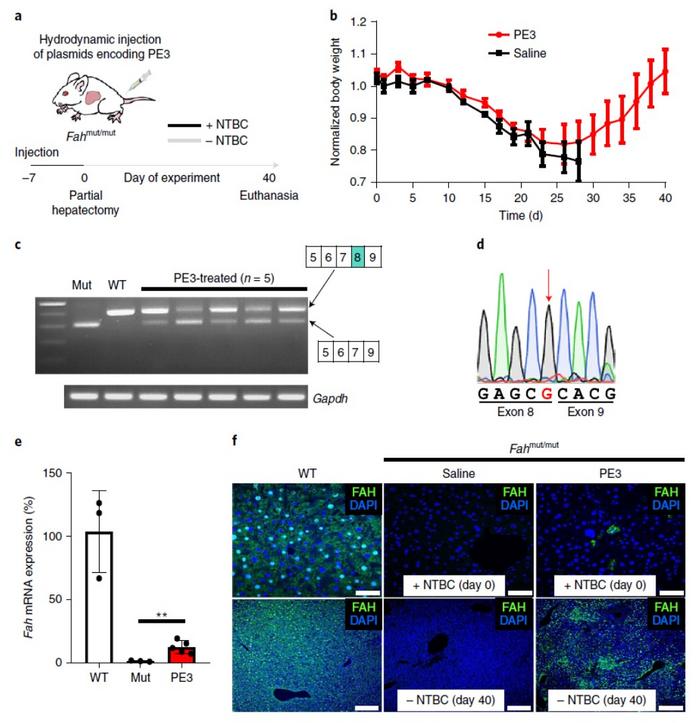
Editing efficiencies, delivery challenges, and off-target effects
In the hereditary tyrosinemia experiments, PE3 editing efficiencies in FAHmut-mut murine liver cells were 11.5% at day 40 post-treatment - comparable to previous gene-editing approaches. The exciting result, however, was the extremely low off-target editing. PE3 treatment resulted in an average off-target indel level of only 0.78% - significantly lower than the 26% achieved by a CRISPR-Cas9 homology directed repair (HDR)-mediated approach.
LCA experiments using the PE2 system proved more challenging in terms of the delivery of the editing components, so the team adopted a different approach.
»The delivery of genome-editing machinery is the major bottleneck in the development of therapeutic genome editing. We used AAV because it is believed to be the safest gene therapy platform. Currently, there are 2 FDA-approved gene therapy drugs, both of which are based on AAV,« Jo explains.
To combat the problem of packaging limit, Jo and the team used trans-splicing AAV (tsAAV) vectors, splitting the two halves of PE2 across two tsAAV vectors, and packaging the pegRNA and sgRNA targeting a mutation in the Atp7B gene in a separate AAV vector. Editing components were then injected into the retina and RPE of the mice, and the resulting editing efficiencies averaged 6.4%.
Importantly, PE2 and PE3 editing in both mouse models of disease showed very little, or no, off-target effects – significantly lower than those seen using other gene-editing technologies.
»Compared to original Cas9 which induces double-strand breaks, prime editing, especially PE2, is a safer strategy in terms of off-target effects. [Our] thorough analyses showed that off-target effects of prime editing were negligible (below 0.1%). Although the off-target effects depend on specific target sequences, current evidence indicates that they are usually low with well-designed prime-editing strategies,« Jo expounds.
The translation of gene therapies is often limited by low in vivo editing efficiencies. Some conditions require very high editing efficiencies in order to be beneficial. For some diseases, positive selection leads to corrected cells expanding and outcompeting mutant cells, meaning that relatively low levels of corrected cells are necessary.
Professor Eric Olson, of the University of Texas Southwestern Medical Centre, a gene therapy researcher who was not involved in the study, suggests that while the paper did not achieve high editing efficiencies, the lack of off-target edits is a key strength:
»Their editing efficiencies in vivo are relatively low, which at least in part reflects challenges with efficient delivery of the prime-editing components. If this low efficiency applies to other targets, the therapeutic applicability will depend on how much of the corrected protein is needed for therapeutic benefit. Some disorders can be rescued with low levels of the missing gene, while others may require physiological levels. Finally, there were few off-target edits, highlighting the potential safety of the method,« Olson observes.
However, Jo asserts that gene-corrected cells selectively expanded in the mouse model of hereditary tyrosinemia, indicating positive selection and meaning that their initial editing efficiencies are more than sufficient.
While there was no positive selection in the RPE of the mouse model of LCA, Jo says this is because most retinal constituent cells are believed to be terminally differentiated. Regardless, previous studies using mouse models of human retinal disease have shown that editing frequencies as low as 1.17% result in significant improvement in retinal function. This fact, combined with the substantial improvement in vision in the PE2-treated mice, suggests that the editing efficiencies achieved by Jo and his colleagues may be adequate.
Comparing different prime-editing systems
The Liu lab’s seminal paper indicates that PE3 and PE3b systems typically have higher editing efficiencies in mammalian cells. However, Jo and the team wanted to test if this is always the case. Because one of the key factors affecting the efficiency of PE2 systems is Cas9 activity, Jo and his colleagues needed to narrow down their search for target sequences on which Cas9 had high predicted activity.
They did so by employing their own deep learning-based model, known as DeepSpCas9. This open-source tool uses a high-throughput approach to evaluate SpCas9 activity at a range of target sequences based on a human cell library containing single-guide RNA and target sequence pairs.
»Current evidence indicates no “better” option of prime editing, but “proper” choice for specific targets,« Jo asserts.
By using DeepSpCas9, the authors hypothesised that they could identify target sequences with high editing activity and, in theory, prove that PE2 can achieve similar efficiencies in mammalian cells as PE3.
A novel screening method to identify the most efficient pegRNAs
One of the most challenging and time-consuming aspects of prime-editing experiments is finding the most efficient prime-editing guide RNAs (pegRNAs) for the targets. In the latest work, Dr. Jo and his colleagues employed an innovative method to screen vast libraries of pegRNAs, which was published in Nature Biotechnology by co-author Professor Hyongbum Henry Kim earlier this year.
This high-throughput screening approach allowed the team to test 435 pegRNAs for hereditary tyrosinemia and 561 pegRNAs for LCA, from which they selected one or two with the highest predicted efficiencies.
»I believe that this high-throughput screening is one of the critical factors of successful in vivo prime editing in this research,« Jo says.
Professor Eric Olson recently used prime editing to correct pathogenic mutations that cause Duchenne muscular dystrophy, and he finds the current study interesting:
»[The authors of the study] describe a clever screen to identify the most optimal pegRNA to correct pathogenic point mutations. Given the dependency of editing efficiency on pegRNA sequence and the inability to accurately predict the optimal pegRNA, this method could be generally useful,« Olson comments.
The importance of teamwork, and moving toward clinical trials
Jo highlights the fact that this study was only possible through the hard work of many team members, including Ms. Hyewon Jang, a dedicated Ph.D. candidate and co-first author. He is also hopeful that the gene-editing field will continue to grow, and that prime editing and other techniques will be able to provide cures for patients with genetic disease in the future.
»I think that the history of CRISPR and CRISPR-related technologies is an excellent example of collective intelligence. Researchers worldwide have solved many issues regarding CRISPR technologies independently but in a coordinated way. Still, there are several issues to be resolved for the successful implementation of genome editing to treat patients with genetic diseases. I hope that more talented researchers will dive into this field of opportunity. Fingers crossed for all the attempts,« Jo remarks.
Jo also notes that the team are gearing up for clinical trials using prime editing to treat these conditions in human subjects.
»Although the funding for research and clinical trials of genome editing is limited, we are preparing investigator-initiated clinical trials using prime editing for patients with paediatric retinal diseases. Hopefully, we will treat the first group of patients in 3 to 5 years,« Jo says.
The innovative methods presented in the paper, along with reasonable editing efficiencies and clear evidence of the safety of prime editing systems in vivo are significant in moving this technology research toward the clinic.
»This is a promising step forward for this new technology, which is moving very fast toward clinical application,« Professor Eric Olson concludes.
Link to the original paper published in Nature Biomedical Engineering:
Rebecca Roberts is a molecular biologist and science writer/communicator based in Queensland, Australia.
Tags
ArticleInterviewNewsIn vivoHydrodynamic deliveryAdeno-associated virus (AAV)Leber Congenital AmaurosisRare DiseaseBase editorsCRISPR-CasPrime editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







