Hyper-Efficient dCas12a Regulates Multiple Genes in Mammalian Retinas
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

The development of CRISPR activation (CRISPRa), CRISPR inhibition (CRISPRi), and epigenome editing (eGE) systems has allowed researchers to regulate the expression of genes rather than creating permanent, heritable edits in the genome. These technologies have opened new doors for therapeutic applications of CRISPR, helping to overcome the hesitancy surrounding permanent editing based on repair of double-stranded breaks (DSBs) in DNA.
In a key development for the field of gene regulation, Lei Stanley Qi’s research group at Stanford University has engineered a new Cas12a protein with expanded capabilities. In an article published this month in Nature Cell Biology, the team describes how the new hyperdCas12a variant can regulate the expression of multiple genes at once with high efficiency, including both gene activation and gene repression. With promising results of multiplexed gene activation in mouse retinas, hyperdCas12a has many exciting potential applications, including therapeutics.
The study is technically robust and multidisciplinary, and is the result of major collaborative efforts from several different departments and institutes at Stanford University. Co-lead author Dr. Lucie Guo says the paper demonstrates the translational potential of the method and highlights the potential for multidisciplinary endeavours:
»The paper is unique because we started from structure-guided protein engineering and went all the way to testing the method in an animal model. It's drawing interest from the CRISPR gene-editing field, as well as from clinical ophthalmologists - I envisioned a way to interface those two worlds.«
HyperdCas12a: A quadruple mutant that packs a punch
CRISPRa and CRISPRi systems use a catalytically dead Cas9 (dCas9), which recognises and binds DNA but does not induce DSBs; by fusing dCas9 to transcriptional effectors, it can regulate the expression of the target gene.
The Qi lab pioneered genome regulation technology and developed the first dCas9 systems for CRISPRa and CRISPRi. However, one drawback of using Cas9 is that it can only target one gene at a time. Enter Cas12a: with intrinsic RNase activity, this protein can process multiple crRNAs from one transcript, offering multiplexed gene targeting.
When Guo first joined Qi’s lab, she tried to adapt the mindset of a clinician, with expectations of using wild-type Cas12a for gene therapy. Despite having a PhD in biochemistry, she never intended to develop a new Cas protein. But necessity is the mother of invention; despite its multiplexing ability, wild-type Cas12a typically achieves lower editing and regulation, and more variable indel efficiencies, drawbacks which have hampered its use in vivo. After abysmal results with the wild-type Cas12 and months of trying to figure out why this was the case, the COVID-19 pandemic hit, driving the team out of the lab.
»That was when we went back to the crystal structure and started looking at the way the Cas12a protein interacts with DNA. We had the thought: if we strengthen the interactions between the protein and the target DNA, we could make this work better. So that was where we started thinking about re-engineering the system,« Guo recalls.
The team started mutating negatively charged residues of aspartate and glutamate to positively charged arginine to increase the binding affinity of dCas12a to DNA. After screening single mutants and identifying those that achieved better gene activation when fused to a miniaturised VPR transcription activator (miniVPR), they began combining them into double, and then triple mutants.
»Ultimately, we arrived at the quadruple mutant, hyperdCas12a, which was achieving the levels of activation we needed. Now we can do the experiments that we could never do with the wild-type version,« Guo explains.
After demonstrating that the quadruple-mutant protein achieved significantly better gene activation results than its wild-type counterpart, the team tested hyperdCas12a for its gene repression activity by fusing it to a KRAB repressor domain, demonstrating that it also achieved better results than its wild-type counterpart.
The power of multiplexing for reprogramming cells and genetic screening
One of the key advantages of the hyperdCas12a system, beyond its significantly higher activation and repression efficiency compared to wild-type dCas12a, is that it can be used for multiplexed genome manipulation. This offers several exciting possibilities for its application, including cellular reprogramming and combinatorial screening.
To demonstrate the power of hyperdCas12a for multiplexed gene regulation, the team first tested it in mouse P19 cells, derived from embryonic teratocarcinoma. The team achieved simultaneous activation of the endogenous transcription factors Sox2, Klf4, and Oct4 using a single crRNA array driven by the well-established U6 promoter, which is commonly used in CRISPR experiments because of its ability to drive expression of sgRNAs. The activation results obtained using hyperdCas12a were significantly higher than the wild-type dCas12a – in the case of Klf4, there was a 70-fold increase in gene activation.
If the Sox2, Klf4, and Oct4 genes sound familiar, it’s because they are 3 of the 4 famous ‘Yamanaka factors’ that are targeted in order to reprogramme adult cells, such as blood or skin cells, into induced pluripotent stem cells (iPSCs). While Guo and her colleagues did not intend to use the hyperdCas12a system to create iPSCs, multiplexed activation of the Yamanaka factors to do so is certainly an intriguing possibility.
Another clear application of hyperdCas12a is in advanced genetic screening. While CRISPR-Cas9 has already revolutionised genetic screening, combinatorial screening - where multiple genes are perturbed simultaneously – remains a challenge. In comparison, the hyperdCas12a system can be used for combinatorial screening with relative ease.
Guo says hyperdCas12a may allow researchers to utilise synergistic relationships between genes, rather than competing to look for ‘silver bullets’ for diseases that are influenced by multiple genes.
»We could use this approach to target several different pathways that all play a role in a very complex disease. There could be a time where these systems-based, multi-pathway approaches become really useful. I hope that toolkits like ours get people at least thinking along those lines,« Guo notes.
While the team used six guide RNAs in a single crRNA array to target three different genes, Guo says this is not necessarily the upper limit of the system, and other avenues are being explored:
»The promoter is the technical detail - the limit ends up being how much the promoter can handle. We used the U6 promoter, which can transcribe around 300 base pairs, so that was the limit, but we are testing other promoters as well.«
The therapeutic potential of gene regulation in vivo
The final test of the hyperdCas12a system was to synergistically co-activate the Sox2, Klf4, and Oct4 genes in vivo in mouse retinas, for which Guo and her colleagues teamed up with Sui Wang’s laboratory at Stanford University's Byers Eye Institute. For Guo, this aspect of the study was particularly important given her clinical training, and it led to exciting results in terms of the regenerative potential of the technique (Figure 1).
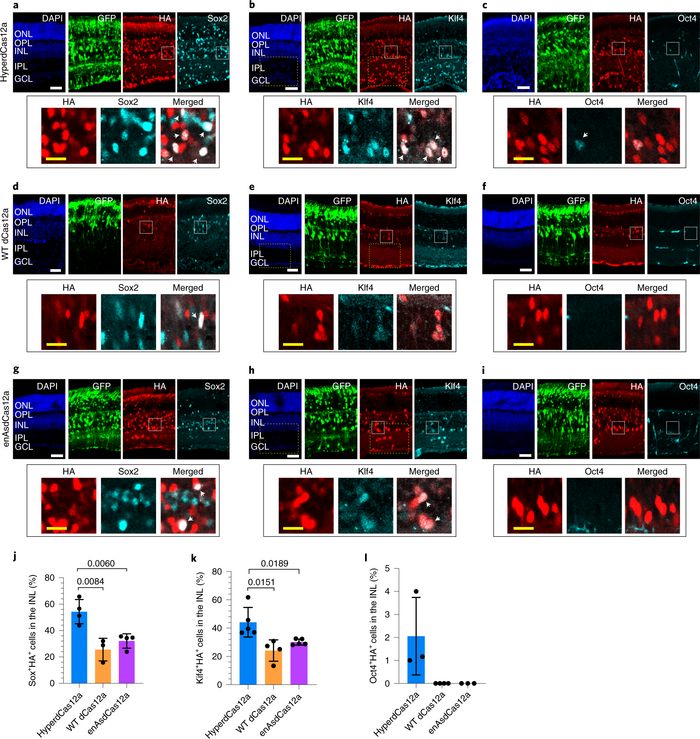
»My interest in ophthalmology, in the retina and blinding diseases of the eye – that’s what drove me to want to take [hyperdCas12a] into an animal model. We wanted to combine the power of gene therapy with how accessible the eye is as a target organ. What we were able to show is that we could upregulate these three transcription factors to drive these retinal progenitor cells to start migrating into the layer of the retina that they don't usually go into,« Guo comments.
Importantly, for the broader gene-editing community, Guo and her colleagues in Yang Hu’s lab, also at Byers Eye Institute, additionally tested the nuclease-active hyperCas12a delivered to mice in vivo via adeno-associated virus (AAV), demonstrating that it was able to perform gene knockout more effectively than its wild-type counterpart.
However, an important aspect of the hyperdCas12a system is that CRISPR technology can be used therapeutically without inducing DSBs in DNA, reducing many of the potential negative consequences. This is particularly relevant considering the increasing number of CRISPR cell and gene therapies that are currently being developed and evaluated in clinical trials, as well as the negative views of permanent gene edits held by many.
Guo and her colleagues are continuing to adapt and improve the hyperdCas12a system, including driving the technology towards clinical applications.
»The next step for this project is to work on applications that are even more disease relevant, and there's a lot of really interesting animal models of retinal disease out there that we will consider,« Guo concludes.
Link to the original paper in Nature Cell Biology:
Multiplexed genome regulation in vivo with hyper-efficient Cas12a.
Rebecca Roberts, Ph.D. is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







