In Vivo Base Editing Corrects Rare Retinal Disease and Restores Sight in Animal Model
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

A recent proof-of-concept study led by Krzysztof Palczewski, Professor in Ophthalmology at University of California Irvine (UCI), supports CRISPR base editing as a new potential treatment modality for Leber congenital amaurosis (LCA), a group of rare and incurable inherited retinal diseases that manifest at birth or in the first few months of life and progress over time, leading to blindness by the 3rd or 4th decade of life.
The recent findings, which were published in Nature Biomedical Engineering late last year, demonstrated for the first time that base editing can correct an LCA-causing mutation in vitro and in vivo, with restoration of visual function in a mouse model for retinal disease.
We spoke with Susie Suh, MD PhD student in Krzysztof Palczewski’s lab at UCI and co-first author on the recent publication to hear more about the work.
»I joined Krzysztof’s lab in 2016, around the same time that base editors began to emerge. I was initially planning to use CRISPR-Cas9 and homology-directed repair to correct RPE65 mutations in a mouse model of retinal disease, but I quickly discovered that the editing efficiency rate was very low with this approach. Others in the field reported similar findings around the same time as well as high levels of off-target edits, so I decided to explore base editing instead.«, explains Susie Suh about how this work, which is essentially her PhD project, started.
Additional Therapies Needed For Leber Congenital Amaurosis (LCA)
LCA arises from loss-of-function mutations in any one of at least 27 known genes that play a role in retinal function, with disease severity depending on the exact mutations present. Mutations in any of these genes result in dysfunctional rods and cones, which are the light-gathering cells of the retina, and mutations in the RPE65 gene are the most prevalent. RPE65 is a critical isomerase in the classical visual cycle in vertebrates that regenerates the active visual chromophore 11-cis-retinal.
LCA is estimated to occur in 1-2 out of 100,000 live births worldwide, and until the recent approval of the gene therapy Luxturna (first approved in the US in 2017), treatment was limited to symptom control and supportive care.
Luxturna, which is based on the sub-retinal administration of a functional RPE65 cDNA delivered via an adeno-associated virus (AAV), represents the first dedicated therapy for LCA. It improves vision but is not curative and concerns exist about its long-term efficacy, with reports of continual retinal degeneration after 1-3 years. Furthermore, its use is restricted to a subset of children and adults that have mutations in both RPE65 alleles and who have a certain threshold of retinal function at the time of treatment.
Although not discussed here, CRISPR-Cas gene-editing technology is also being pursued as a therapeutic strategy for LCA, and the first patients were treated withEditas Medicine’s EDIT-101 last March. EDIT-101 is based on CRISPR-Cas correction of mutations in the CEP290 gene, which are responsible for LCA10, which accounts for 20-30 % of all LCA cases.
Potential in Base Editors
Base editors, which were first developed in David Liu’s lab at Harvard University in 2016 are essentially an RNA-guided Cas protein domain fused to a deaminase domain that can convert one nucleotide into another and thereby introduce or correct single point mutations (see Fact Box). Base editing has been investigated for various animal disease models, but Krzysztof Palczewski’s group is the first to use the technology to correct LCA-associated mutations.
Given that base editors can ‘rewrite’ single bases without the requirement for double-stranded DNA break (DSB) formation, they are attractive in therapeutic applications from a safety point of view. Attractive as they may be, however, base editors can be challenging to work with, as Susie Suh explains:
»Their activity is reliant on a number of stringent criteria being met. Besides the obvious need for the base-editing enzyme itself and a target-specific guide RNA (gRNA), the editing strategy must also take into account the activity window of the base editor.«
The activity window is the region within the genome that is typically defined by the number of nucleotides from the protospacer adjacent motif (PAM), in which a given base editor can induce efficient point mutations. The activity window for most base editors is approximately four to five nucleotides wide.
The small size of the activity window means that in practice, the range of sequences that can be targeted by a base editor is narrow, as Susie Suh continues: »The base that you want to correct should be within a four to eight base pair window where the gRNA binds, and the gRNA should be targeted with the NGG PAM sequence.«
Just as engineered Cas variants with less stringent PAM requirements have emerged, base editors that can recognise a broader range of PAM sequences have also been developed. Susie Suh and her co-workers took advantage of these variants to correct a nonsense mutation in the RPE65 gene on exon 3 in vitro, and later in the rd12 mouse model, which is an established disease model for human LCA.
FACT BOX: Base Editors
Two types of base editors have been developed so far: cytosine base editors (CBEs) and adenine base editors (ABEs), which are fusions of a catalytically-dead Cas9 (dCas9) fused to either a cytidine deaminase or an adenosine deaminase. When introduced into a cell, CBEs and ABEs enable the conversion of C·G to T·A or A·T to G·C, respectively, with high precision and efficiency.
The major advantages of base editors over CRISPR-Cas9 homology-directed repair (HDR) is that they can perform targeted and precise conversion of a single base pair independently of HDR and double strand breaks (DSBs), which allows for precise and efficient gene editing without the safety risks associated with DSBs, and without the need for template-based HDR.
Base editors are gaining traction within the gene therapy field for the precise correction of various genetic disorders.
In Vitro Screening of RPE65 Base Editing
Using an rd12 reporter cell line that was engineered to express mutant RPE65 corresponding to the rd12 mouse, the researchers tested the efficiency of various adenine base editors (ABEs) in combination with a set of target-specific gRNAs.
Correction of the nonsense mutation should restore RPE65 expression, which could easily be detected by western blot analysis. Deep sequencing confirmed the base correction and allowed quantification of the conversion of adenine to guanine at the target site in RPE65.
In vitro, the highest base editing efficiency rates were observed when the codon-optimised ABEmax was used with either of two gRNAs (gRNA-5 or -6.) Off-target edits outside of the base-editing window were below background levels compared to untreated RPE tissue, in line with other reports about low off-target rates for ABEs.
Restored Retinal and Visual Function
Before treatment, the rd12 mutation abolishes RPE65 expression in mice, resulting in markedly reduced sight and a blockade in the visual cycle due to a lack of 11-cis-retinal. Conveniently, 11-cis-retinal levels can be measured as a direct biochemical readout for an intact visual cycle.
Measurement of 11-cis-retinal levels by high-performance liquid chromatography revealed that eyes from rd12 mice treated with the ABEmax and gRNA-5 had substantial 11-cis-retinal levels, and that the restored supply responded to flash stimuli as expected for a healthy visual cycle.
Further assessment of rd12 mice by a series of visual function tests revealed that the eyes of base-edited mice behaved comparably to healthy eyes in terms of photoreceptor function and downstream retinal interneurons in response to flash stimuli and that vision was restored substantially.
Base Editing Corrects RPE65 Mutation in Mouse LCA Model
With success in vitro, the researchers then proceeded to test their base-editing strategy in rd12 mice. To this end, they made a set of lenteviral vectors that express ABEmax along with either a target-specific or a control gRNA.
Since this was a proof-of-concept study, the researchers chose lentiviral over the safer AVV delivery method for two main reasons. Firstly, lentivirus enables delivery of a transgene that encodes the base editor and gRNA owing to its large gene transfer capacity, while AAVs can only accommodate transgenes that are <4.7 kb. Secondly, lentevirus has a natural tropism for the retinal pigment epithelium (RPE), thus circumventing the need for a tissue-specific promoter and maximising the likelihood of efficient base editing. For Susie Suh, in vivo administration of the test treatment was the biggest challenge in this study:
»Sub-retinal injection into mice is one of the most challenging techniques used in the paper, because it’s difficult to inject virus right between the retina and RPE tissue, while using the microscope. Ideally, we wanted to transduce at least 70% of the RPE cells with the virus without damaging the mouse eye for functional assessment. Unlike intraperitoneal injection or oral gavage, it requires a large amount of effort in sub-retinal injection and screening to obtain the treated samples for the study.«
Despite the challenge, the researchers succeeded, with restored and correctly localised RPE65 expression in the eyes of rd12 mice 5 weeks after injection.
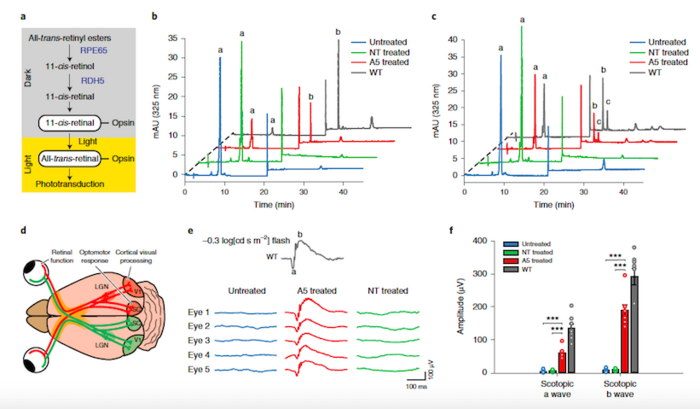
A New Curative Treatment Modality for LCA
These new findings mark the first proof-of-concept for base editing as a treatment modality for LCA and according to Susie Suh, the RPE65 rescue rates seen in rd12 mice are very encouraging: »In our mouse model, we saw significant restoration of visual function with only 30 % base correction. In fact, in humans and mice, carriers of loss-of-function RPE65 variants have visual function that is equivalent to that of non-carriers. This shows how robust the visual cycle is and how well it might respond to even partial restoration and we expect the situation to be similar in humans.«
However, before we can expect to see preclinical development the researchers have a few open questions to address. Besides optimising the editing efficiency further, they are also exploring whether or not their base-editing strategy can rescue visual function in older rd12 mice, taking into account knowledge from the LCA field that it becomes difficult to treat the disease when it has progressed too far. They will also look into additional clinical phenotypes in the rd12 mice, such as the loss of colour vision that is associated with LCA, to get a deeper understanding of the consequences and therapeutic outcomes of base editing.
Jeong Hun Kim, Professor in Biomedical Sciences at the Seoul National University College of Medicine (South Korea) who has explored CRISPR-Cas9-mediated correction of the same nonsense mutation in Rpe65 in rd12 mice, but who is not involved in the current work sees great potential in using base editors to cure LCA:
»As a clinician scientist within genome editing for retinal diseases, I am interested in CRISPR-Cas9, base editing and prime editing. About 75% of likely pathogenic or pathogenic variants of the RPE65 gene are substitutions according to the ClinVar database. Base editing and in particular, adenine base editing, can correct these mutations. Genome editing including base editing might be a permanent and direct approach to recover wild-type sequences at the endogenous locus, unlike exogenous gene therapy. The impact of base editing might be increased with the improved versions of base editors, which are being reported, in terms of specificity and efficacy.«
A Platform To Correct all LCA Mutations
The study described here aims to correct a specific mutation in RPE65, but in the long term, the researchers hope to use base editing as a platform to correct all LCA mutations. While additional development programs may emerge for other types of LCA based on specific mutations, Susie Suh and her colleagues hope that in the long-term, base editing can be used to correct all of the known LCA mutations:
»Here, we have shown that it’s possible to correct one specific sequence with base editing, but we see the real potential in developing a platform to screen for the perfect pair of base editor and gRNA that can be applied to correct any target sequence.«
Link to original article (and source of figures used in this article) in Nature Biomedical Engineering:
Tags
ArticleInterviewNewsIn vivoLentivirus (LV)Leber Congenital AmaurosisGene therapyBase editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







