Researchers engineer programmable gene activation cascades
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
The study builds on a broader effort to make gene-regulation tools behave less like static switches and more like the developmental programs they aim to emulate. Traditional CRISPR activators can raise or lower gene expression, but they do so simultaneously, lacking the temporal choreography that cells rely on during embryogenesis and tissue formation. A team at Syntax Bio set out to build a system that could impose this missing dimension: not merely which genes are expressed, but when, and in what order.
“The key missing piece of the puzzle was the ability to also encode time, which is what the proGuide system enables”Ryan Clarke, Syntax Bio
At the centre of the study lies a long-standing challenge: CRISPR-based activators offer synthetic control over gene expression, but they typically lack temporal structure. And timing really matters, Ryan Clarke underscores. He is cofounder and CTO of Syntax Bio, which developed the new genetic system, and he is also co-senior author of the paper presenting it in Science Advances.
»Transcription factors are the epicentre of controlling what cell type a stem cell becomes in human development. If you have a synthetic transcription factor, like CRISPRa/CRISPRi, that's a powerful starting point for programming and engineering these processes. The key missing piece of the puzzle was the ability to also encode time, which is what the proGuide system enables – precise time-ordering of the expression of the key genes that drive development for a specific cell type,« he explains.
The mechanism hinges on conditional guide RNAs carrying a Pol III terminator that prevents their transcription (see Figure 1). Only when a preceding guide directs Cas9 to remove this terminator does the next guide become active. In this study, Cas9 is fused to VPR, a three-part activation domain (VP64–p65–Rta) that converts nuclease-dead Cas9 into a strong CRISPRa effector. Cas9-VPR therefore drives the system in two ways: it cuts proGuide plasmids to convert them into active guides, and it activates endogenous target genes once those guides are produced.
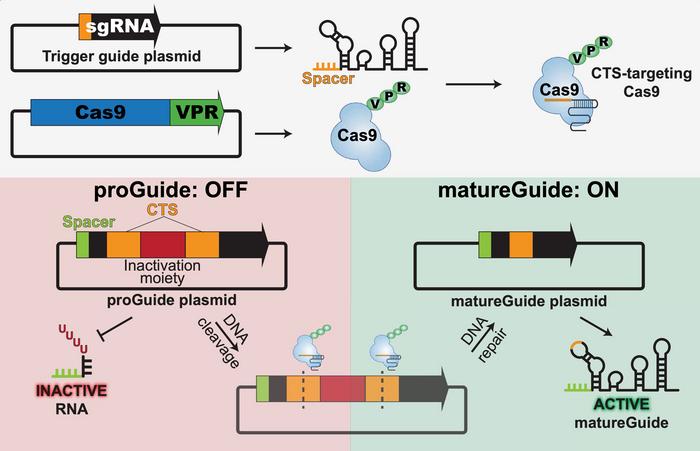
Each proGuide carries not only its own spacer sequence but also a 23-bp Cas9 target site (CTS) embedded within its DNA. This CTS serves as the recognition sequence for the previous guide in the cascade. When that earlier guide becomes active, it binds the CTS and directs Cas9-VPR to cut at that location, excising the Pol III terminator and thereby converting the downstream proGuide into a functional matureGuide. In other words, the CTS provides the address label that determines which proGuide gets activated next, allowing the system to link individual guide RNAs into a programmed, stepwise chain.
Early versions of the design suffered from polymerase read-through, but the authors resolved this by extending the poly-T tract and introducing a second terminator, separated by a short linear segment, which suppressed background activity to almost undetectable levels. They then refined how Cas9 excises the terminator by arranging cut sites as inverted repeats, producing clean deletions and consistently functional matureGuides.
To ensure that each cascade step would fire reliably, the team computationally generated millions of potential Cas9 target sequences and experimentally winnowed them to a panel with strong, low-leak performance. Test cascades showed the expected gradual loss of efficiency with increasing step number but no catastrophic failure, indicating that the optimised parts could support deep sequential logic.
“What we do is different. We’re giving them the instructions from the inside, and then they read that first set of instructions, conform, go to the next step, conform”Brad Merrill, Syntax Bio
With this architecture, the authors built CRISPR activation cascades that triggered endogenous gene expression in an ordered manner. In HEK293T cells, sequential activation of CXCR4 unfolded with a reproducible lag between steps, and multi-gene circuits exhibited activation patterns determined by wiring rather than intrinsic promoter behaviour.
The real stress test came in human iPSCs (see Figure 2). Here, cascades directing DLL4, CD4 and CD105 demonstrated robust position-dependent activation: genes placed earlier in the circuit emerged sooner and in more cells, even though all three markers typically show low baseline expression. The strategy recalls a broader critique of conventional differentiation methods.
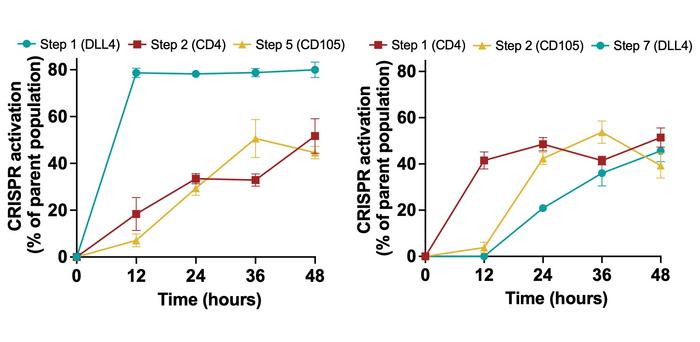
»Traditionally, you build a tissue culture environment, and then cells enter it and will eventually conform to that environment over time. Then, in order to get them to the next stage, you need to build a different environment,« explains Brad Merrill. He is a cofounder and Head of Innovation at Syntax Bio, and a co-senior author of the Science Advances paper.
Because those environments involve 'infinite variables', he argues, the field has struggled to achieve predictable outcomes. The proGuide system attempts something more direct, Brad Merrill points out:
»What we do is different. We’re giving them the instructions from the inside, and then they read that first set of instructions, conform, go to the next step, conform.«
That perspective reflects the work's founding motivation. Merrill says the team began with the view that, despite years of effort, there may be an underlying weakness in the approach to stem-cell differentiation, and that programmable intracellular instruction sets might bypass that weakness to improve the reliability of producing specific cell types.
Although questions remain about how to align cascade pacing with cellular rhythms or extend activity over longer developmental windows, the study demonstrates a practical means of encoding temporal logic directly into cells. By allowing CRISPR to follow developmental syntax rather than forcing abrupt leaps, the system provides a controlled framework for interrogating – and potentially improving – the transitions that underlie engineered cell fates.
Co-first authors of the study were Anupama Puppala and Andrew Nielsen, while Ryan Clarke and Bradley Merrill were co-senior authors – all at Syntax Bio Inc. in Chicago, Illinois. The research was published in Science Advances on 5 December 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







