Safety: CRISPR cause virus vector to integrate at high level. Interview: Bence György
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
(Interview is condensed and edited for clarity)

- Virus vectors integrating into host DNA sounds very bad as in risk of causing cancer, how concerned should we be, is this going to end AAV's as vectors for CRISPR gene therapy?
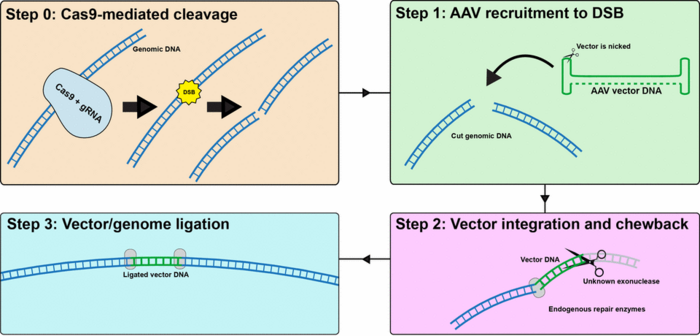
No no no, we are not saying we can't use AAV for CRISPR delivery. In fact, our data suggests that at least in the rodent system expression of Cas9 and gRNAs does not increase AAV integration. AAV vectors are known to integrate at a low rate (~0.1%) of incoming vector at DNA breaks. Since CRISPR makes DNA breaks, in hindsight, it isn’t surprising that AAV also integrates into these cut sites. The important observation that we made is that this basal level of integration is not enhanced with CRISPR, and that the very specific cutting with well-designed guides just provides AAV with another potential integration site. At this point we have to characterize the AAV CRISPR system fully and make sure we know that integration is happening and detect it with specific protocols.
AAV integration is very common
- ok, lets take a few steps back, can you explain a bit about the background for the paper?
Yes. We previously published a Nature Medicine paper on a dominant form of hearing loss. We used AAV CRISPR to make a doublestranded break and knock out the function of dominant deafness gene, Tmc1, in inner-ear hair cells.
We had mice that were completely deaf and after AAV CRISPR injection they were normal hearing up to one year after. It was really astonishingly.
High levels of AAV vector integration into CRISPR-induced DNA breaks
The new study 'High levels of AAV vector integration into CRISPR-induced DNA breaks' was published in Nature Communications by Bence György, MD, PhD with collaborators Killian Hanlon, PhD and Casey Maguire, PhD at the Massachusetts General Hospital and Harvard Medical School.
- but then You also observed AAV integration or?
Yes and we wanted to create a AAV based platform for CRISPR delivery into cells so we were wondering what actually happened in the cells. Doing sequencing we observed the virus went into the gene we were targeting - essentially it was AAV integration - we really did not expect the AAV genome to be there, although in hindsight, it is not very surprising since it is well known that AAV integrates at a low frequency at DNA breaks.
“in some cases every second cell would have an AAV in the targeted gene”
We observed that this was a very common outcome of CRISPR applications - in some cases every second cell would have an AAV in the targeted gene.
We looked at genes in the brain, the ear, and in muscles because these are also therapeutic targets for CRISPR and all the time we saw AAV integration if CRISPR was present.
Safety of AAV integration?
- Ok, but the good news is that You found it only to integrate in the CRISPR target gene?
As mentioned earlier, AAV can already integrate at a low frequency throughout the genome. This has not been associated with any pathology in clinical trials to date. Our data shows that the specific CRISPR cut site gives AAV another break for potential integration.
First of all we showed that CRISPR did not cause an increase in genome-wide integration of AAV, but only at the CRISPR-cut site.
Second we used a very short AAV vector to be able to sequence the entire integrated region in one piece and really study the mechanism of integration.
Doing that we found the majority of AAV gets in fragmented but we also observe full length AAV.
Whether it affects the safety is a good question.
- yes?
Since AAV already integrates, and CRISPR is only adding one additional site, it does not obviously impact safety concerns over what is already occuring with AAV vectors in the first place.
However, if poor guides (non-specific) guides were designed, one could imagine more off-target cuts would be made and this could increase AAV integration over basal levels. So it is important to use the most efficient Cas9/gRNA system for your target.
Additionally, one should consider where the cut site is in the context of other genes/oncogenes. Integration of the CMV-promoter driven Cas9 could potentially activate genes you don’t want to activate. However, this has not been reported in any clinical trials with AAV vectors which already integrate on their own.
AAV normally safe non-integrating vector
- Ok, before continuing maybe you can explain a bit why you use AAV vector, what makes it a good vector?
Yes. First of all the AAV or Adeno-Associated Virus is a very safe non-pathogenic virus that has been accepted by the FDA as a therapy for two diseases so far, retinal therapy for blindness, and spinal muscular atrophy to treat a horrific disease in infants. It is very effective and currently there is no better delivery tool for nondividing tissue like brain, muscle, eye or ear.
- but being in non-dividing cells also means it will stay and express the CRISPR Cas9 for a long time?
Correct, this is a concern. In dogs AAV encoded transgenes have been shown that expression can be there for a decade. So this is a concern because it is a long term expression and there is a risk because something might happen even a decade later (like immunogenicity to Cas9 or off target cutting).
Of course there are many questions. You have to ask, what will happen? Formation of a tumor or may be an immune reaction at some point or degeneration of the target cells?
Basically we now know the integration is happening already with AAV vectors and we have to look at the safety aspects and analyze this specifically. From our studies we never saw hard outcomes like increase in mortality, cancer formation or genotoxicity.
Developments could adress concerns
-what would you point to as the main concern?
The main concern is having an active nuclease there for a long time where it can keep cutting the genome. I think there are several developments in the field can get around it.
- Such as?
You can limit the expression of the nuclease fx make a self-inactivating vector or if you just deliver the CRISPR Cas proteins then it doesn't stay that long. Another option might be not to use an active nuclease, but use the prime editing concept which essentially is using the Cas9 to target, but not to cut the genome. We would like to think about strategies to stop the expression of CRISPR so it is not active very long.
Tool to detect off-target effects
- Ok, also in the paper you suggest the AAV integration could be a sensitive tool to detect genome breaks?
Yes this is also very important since it's very hard to find genome breaks that happen at low frequency. We and others are developing assays looking at AAV integration as way to assay CRISPR specificity and double-stranded breaks across the genome.
- how would this be done?
Instead of looking at the doublestranded breaks themselves we take genomic DNA and PCR the AAV out from the genome. Then you will see genomic fragments as well and can map where the AAV has integrated. We have preliminary data in vitro that looks very nice, so we can detect off-target effects using AAV.
- Great, thank You very much for taking time to talk to us.
Bence György
Bence György, head of the clinical translational research group at the Institute of Molecular and Clinical Ophtamology Basel (IOB), Switzerland, where he is in charge of developing gene editing based therapies towards treatments for genetic forms of blindness using CRISPR Cas9 and AAV vectors.
Tags
ArticleInterviewDeliveryViralAdeno-associated virus (AAV)CRISPR-CasSafety
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







