Highlight: HiFiCas9 Selectively Disables Mutant KRAS in Lung Tumours
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
“Using HiFi-Cas9, we designed guides that specifically recognise the two most common KRAS mutations in lung cancer, cutting only the mutant DNA while leaving the healthy copy intact”Pedro Medina
KRAS mutations at codon 12 represent critical oncogenic drivers in lung adenocarcinoma, occurring in approximately 32% of cases. The KRASG12C (G12C) and KRASG12D (G12D) substitutions arise from single-nucleotide changes that severely impair GTPase activity, leading to constitutive activation of downstream MAPK and PI3K/AKT signalling pathways. This hyperactivation drives uncontrolled cell proliferation and represents a paradigm of oncogene addiction, where tumour cells become dependent on sustained mutant KRAS expression for survival.
The research team employed HiFiCas9, a high-fidelity SpCas9 variant containing the R691A substitution that maintains robust on-target activity whilst reducing off-target cleavage. They designed mutation-specific single guide RNAs (sgRNAs) exploiting distinct protospacer adjacent motif (PAM) sites, positioning the mutated nucleotide within the sgRNA seed region whilst introducing artificial mismatches to enhance specificity against wild-type sequences (see Figure 1).
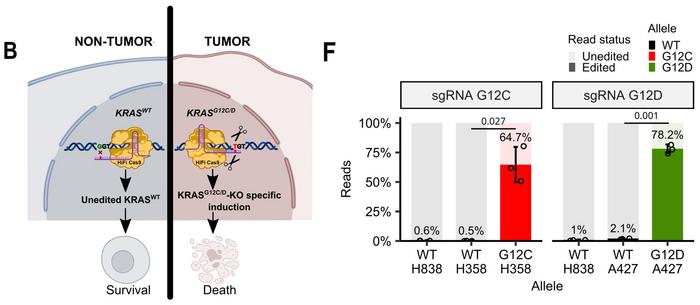
Comprehensive specificity validation demonstrated selective editing only in cell lines harbouring the corresponding mutations, with no detectable activity against wild-type KRAS(KRASWT) alleles. Next-generation sequencing confirmed mutation-specific targeting, revealing editing frequencies of 64.7% and 78.2% for G12C and G12D, respectively, while KRASWT alleles showed editing frequencies below 2.1% (see Figure 1F).
»Instead of blocking the mutant protein, we aimed to eliminate the mutation directly at the DNA level. Using HiFi-Cas9, we designed guides that specifically recognise the two most common KRAS mutations in lung cancer, cutting only the mutant DNA while leaving the healthy copy intact,« Pedro Medina explains to CRISPR Medicine News. He is professor at the University of Granada and senior author of the paper that was published Monday in Nature Communications.
Functional validation revealed significant cytotoxic effects in KRAS-mutant cell lines. Ribonucleoprotein delivery produced viability reductions of 67% (H358, G12C), 26% (H1792, G12C), 36% (A427, G12D) and 32% (SK-LU-1, G12D) at seven days post-treatment, whilst KRASWT H838 cells showed no significant reduction. Effects were enhanced in three-dimensional spheroid cultures, consistent with increased KRAS dependency in physiologically relevant conditions.
“In preclinical models, HiFi-Cas9 slowed tumour growth and even outperformed sotorasib in some cases, including resistant tumours”Pedro Medina
Western blot analysis demonstrated substantial reductions in total KRAS protein levels following HiFiCas9 treatment, with complete knock-out of G12D protein in A427 cells. Downstream signalling analysis revealed decreased phosphorylation of ERK1/2 and p70S6K1, indicating effective disruption of both MAPK and mTOR pathways.
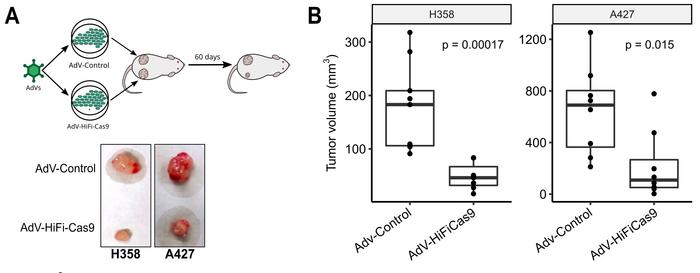
In vivo validation employed both cell-derived xenograft (CDX) and patient-derived xenograft (PDX) models. CDX studies revealed substantial growth inhibition: 63% reduction in H358 tumours and 42% reduction in A427 tumours after 60 days compared to controls (see Figure 2). PDX experiments using direct intratumoral injection demonstrated statistically significant decreases in growth rate for both G12C and G12D models over 28 days.
Comparative analysis with sotorasib, the FDA-approved G12C covalent inhibitor, revealed superior HiFiCas9 performance. Two-way ANOVA analysis indicated that HiFiCas9 accounted for 72.96% of total variance, whilst sotorasib contributed only 13.3%. Combination treatment provided no additional benefit, consistent with non-overlapping mechanisms – genetic elimination versus pharmacological inhibition of GDP-bound protein.
Patient-derived xenograft organoid studies using three models revealed heterogeneous responses reflecting clinical diversity. TP79 responded exclusively to HiFiCas9, with no sensitivity to sotorasib, while TP60 responded to both treatments.
These findings highlight potential therapeutic advantages in sotorasib-refractory cases where high GTP-bound G12C levels may drive resistance.
»In preclinical models, HiFi-Cas9 slowed tumour growth and even outperformed sotorasib in some cases, including resistant tumours. This suggests that CRISPR-based strategies can address vulnerabilities beyond the reach of current drugs and opens the way towards more durable therapies once in vivo delivery is optimised,« Pedro Medina says.
Evaluation of sotorasib-resistant cell lines demonstrated context-dependent efficacy. H23 resistant cells, harbouring co-occurring KEAP1 and SMARCA4 mutations associated with poor clinical outcomes, showed significant viability reduction (60%) in 3D cultures. Sequential treatment experiments revealed no evidence of adaptive resistance development across three consecutive treatments, suggesting durable therapeutic potential.
The study demonstrates that carefully engineered HiFiCas9 systems can achieve clinically relevant single-nucleotide discrimination for oncogene targeting. The superior performance compared to current G12C inhibitors, combined with activity against G12D (for which no approved therapies exist), establishes this approach as a potentially transformative therapeutic strategy requiring advances in in vivo delivery optimisation.
This study was conducted by Pedro Medina, Juan Carlos Álvarez-Pérez, Juan Sanjuán-Hidalgo, Alberto M. Arenas and colleagues from the University of Granada, Spain, in collaboration with collaborators from multiple Spanish institutions. The study was published in Nature Communications on 1 September 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleCMN HighlightsNewsStickyRibonucleoprotein (RNP)Adenovirus (AV)Lung CancerCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







