In Vivo Prime Editing in the Mouse Liver Suggests Therapy for PKU
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Gerald Schwank has been riding the CRISPR train from the very beginning. Already as a postdoc in 2013, he demonstrated functional repair of the gene responsible for cystic fibrosis by gene editing in patient-derived organoids. He is now a professor and heads a research group at the University of Zürich, and the primary goal is the clinical translation of genome-editing tools. In line with this ambition, Gerald Schwank's lab recently published a paper in Science Translational Medicine that demonstrates proof-of-concept for the feasibility of in vivo prime editing to treat mice with the hereditary liver disease phenylketonuria (PKU).
PKU results from one of several hundred inactivating mutations in the phenylalanine hydroxylase (Pah) gene that catalyses the conversion of the amino acid phenylalanine to tyrosine in the liver. Without a functional PAH enzyme, phenylalanine (L-Phe) accumulates to toxic levels in the body, leading to intellectual disability, seizures, behavioural problems, and mental disorders.
The number of people affected by PKU varies considerably from 1:2,600 in Turkey to 1:200,000 in Finland, with the UK and USA having intermediate frequencies of 1:10,000 and 1:25,000, respectively. Currently, there is no cure for the disease and people are treated with a diet low in phenylalanine or, to a lesser extent, with medication that lowers blood levels of the amino acid.
“This study is definitely a stepping stone towards the clinical application of prime editing for treating genetic diseases”Dong Hyun Jo
While gene editing seems like an obvious therapeutic path to follow for this genetic disease, most cases of PKU are caused by point mutations that are not ideally suited for correction by classic CRISPR-Cas9. This is because the method relies on the generation of DNA double-strand breaks and subsequent repair, making it challenging to install precise edits.
Base editing is more precise but is restricted to certain base substitutions and can thus only be used for a subset of all the disease-causing mutations seen in PKU. On the other hand, prime editing is both a versatile and reliable gene-editing technique.
Although prime editing is less efficient than base editing, Gerald Schwank used this strategy in the current study. Dong Hyun Jo at the Seoul National University College of Medicine, Republic of Korea, acknowledges this choice. He was not part of the study but has extensive experience with in vivo prime and base editing in mice with liver and eye diseases. He says:
»Because there are various mutations in the PAH gene linked to PKU, prime editing might be a more plausible treatment option than base editors or original Cas nucleases. In addition, there are fewer concerns about bystander edits - as you see with base editors - and off-target effects by double-stranded breaks - commonly observed with original Cas nucleases - in prime editing.«
Prime editing
Prime editing is a 'search-and-replace' genome-editing technology in molecular biology by which the genome of living organisms may be modified. The technology directly writes new genetic information into a targeted DNA site. It uses a fusion protein, consisting of a catalytically-impaired Cas9 endonuclease fused to an engineered reverse transcriptase enzyme, and a prime-editing guide RNA (pegRNA) that is capable of identifying the target site and providing the new genetic information to replace the target DNA nucleotides. It mediates targeted insertions, deletions, and base-to-base conversions without the need for double-strand breaks (DSBs) or donor DNA templates. From Wikipedia. Read more about prime editing here on CRISPR Medicine News.
Prime editing achieved clinically relevant levels
A mouse model of PKU harbouring the homozygous Pahenu2 locus was used in the study (Figure 1). The locus carries a T>C point mutation on exon 7 of Pah, leading to negligible PAH activity and abnormally high L-Phe concentrations in the blood. Gerald Schwank and his team designed a prime editor guide RNA (pegRNA) that could correct the mutation, and it was delivered in vivo along with the prime editor (PE) in an adeno-associated virus (AAV) or adenovirus (AdV) vector.

After several rounds of optimisation, the team managed to achieve editing rates of 11.1% after injection of the gene-editing cargo into newborn mice. This increased PAH activity from virtually 0% to 6% of wild-type activity and lowered blood L-Phe from around 2000 µmol/l to 100 µmol/l, which is well below the therapeutic threshold of 360 µmol/l.
»A few studies have applied prime editing in vivo before, but they have reached very low editing rates - several-fold lower than us - and therefore diseases like PKU could not have been corrected with previous approaches,« says Gerald Schwank. »We have optimised the editors and the delivery vehicles to increase the efficiency, and now we actually reach efficiencies where we can think of curing many genetic diseases.«
Dong Hyun Jo shares this optimistic view, but he cautions that further optimisation is needed:
»This study is definitely a stepping stone for the clinical application of prime editing for treating genetic diseases. However, issues regarding the delivery and editing efficiencies should be appropriately addressed,« he says.
The successful results did not come easily, and Gerald Schwank's team had to optimise several process steps. These included engineering the PE fusion enzyme, choosing the suitable viral vector, selecting the animals to be treated, and introducing an extra guide RNA called PE3.
Multiple optimisations led to sustained and functional prime editing in the liver
PEs are large enzymes that require more than 6.6 kb of DNA to be encoded, and this already exceeds the packaging capacity of ~4.7 kb for AAV. Therefore, as a first optimisation step, the 0,6 kb RNaseH domain of the fusion enzyme was deleted to create PE2.
“We now work on vectors where we can express the editor at higher levels, and in addition, we try to find a prime editor variant that is re-engineered to be more efficient. Hopefully, these two approaches will allow us to treat mice - and later humans - with lower vector doses”Gerald Schwank
Using PE2ΔRnH, all necessary components could now be encoded in a 7.5 kb construct, which is suitable for AdV delivery but exceeds the carrying capacity of AAV. Gerald Schwank, therefore, used the intein-split technology that allows one to divide a construct into two pieces of around 4.5 kb each, which can be delivered separately by two AAVs. In the cells, the two parts of the encoded protein are subsequently assembled to a functional PE2ΔRnH. A third optimisation step introduced an additional sgRNA called PE3. This causes the PE complex to nick the unedited strand and leads to a more efficient repair of the edited strand.
In vivo PE2ΔRnH prime editing efficiencies of the Pahenu2 locus in the liver of either newborn or adult mice were tested with all combinations of the optimisation steps. Results showed superior performance of AdV compared to AAV in converting the disease-causing C-nucleotide back into the wild-type T-nucleotide. Editing was also several-fold more efficient in neonates than adults, and even better results were obtained when the sgRNA was included with the pegRNA. The best combination - AdV-delivery to neonates including sgRNA - led to a prime editing efficiency of 11.1% (Figure 2).
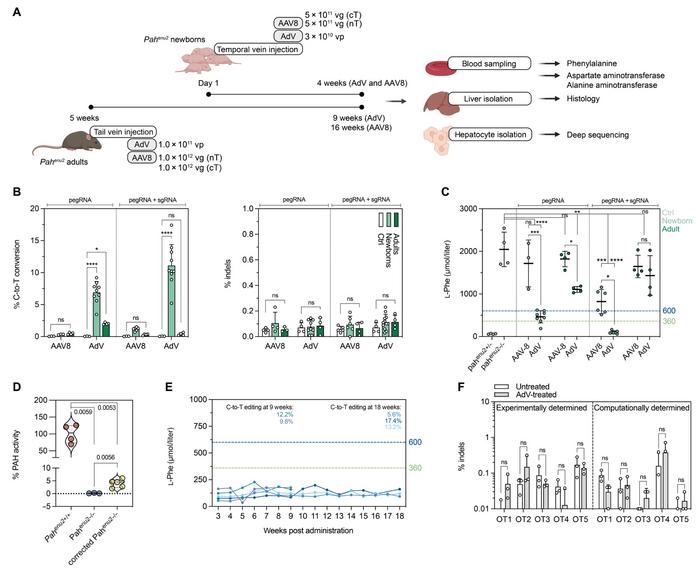
Across all combinations, unintended insertions or deletions (indels) were detected at comparable low levels of around 0.1%. In addition, deep sequencing of 14 different tissues revealed that editing was negligible at approximately 0.05% in all tissues except skeletal muscle, where it reached 0.2% - much less than the 11.1% achieved in the liver. This high tissue specificity was due to hepatotropism of both the type 5 AdV used in the study and the P3 promotor driving expression of PE2ΔRnH.
Editing efficiencies broadly reflected blood L-Phe concentrations after in vivo prime editing. Only AdV-delivery in newborn mice reached therapeutically satisfactory levels (600 and 300 µmol/l in the USA and Europe). These were 4-fold lower in mice treated with sgRNA plus pegRNA than pegRNA only. This latter group of animals displayed between 2% and 6% of wild-type PAH enzyme activity. Both editing rates and L-Phe levels were sustained for at least 18 weeks.
Lower vector doses are required for clinical translation
A general concern with prime editing and gene editing is the risk of toxicity or immune responses induced by the enzymes or viral vectors. Gerald Schwank performed several analyses to address these critical questions. Results showed transiently elevated transaminase alanine aminotransferase (ALT) concentrations that were presumably triggered by mild hepatotoxicity of the liver induced by AAV and AdV.
Moreover, AdV but not AAV induced transient elevation of various inflammatory cytokines and chemokines in the serum and liver. In addition, sustained, elevated liver-resident neutrophils and monocyte-derived macrophages were observed in AdV-treated animals. At the endpoint, animals treated with AdV as adults had also developed specific antibodies to Cas9.
»If we inject high enough doses of the AdV vector, we get sufficient editing rates to cure the PKU phenotype. However, these vector doses are too high for the clinic and will presumably be problematic in humans. So, we must get a more efficient prime-editing enzyme to apply this therapeutic strategy in the clinic,« Gerald Schwank explains.
»We now work on vectors where we can express the editor at higher levels, and in addition, we try to find a prime editor variant that is re-engineered to be more efficient. Hopefully, these two approaches will allow us to treat mice - and later humans - with lower vector doses,« he concludes.
Link to the original article in Science Translational Medicine:
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsDeliveryIn vivoAdeno-associated virus (AAV)Adenovirus (AV)DiseasePhenylketonuria (PKU)Prime editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







