Miniscule Cas nucleases do a Mammoth’s job
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...

Hi Lucas, nice meeting you and having this opportunity to speak with you.
Thanks for reaching out, Gorm.
Maybe you should start by telling a little about yourself and how you got involved in Mammoth?
Mammoth Biosciences is almost four years old now, and we are around 120 people. I founded the company together with Janice Chen, Trevor Martin and Jennifer Doudna. Janice and I were doing our PhD work in Jennifer's lab at the University of California, Berkeley, and we were finding a lot of these new compact CRISPR systems. We were trying to understand some of their properties and biochemistries, and we got motivated to let our discoveries go beyond a paper and see these things translated.
What was the scientific foundation for the new company?
We started Mammoth at the beginning of 2018, initially focusing on CRISPR diagnostics. We had developed our DETECTR type V trans cleavage system to diagnose COVID-19, but the premise of the company was to use the diversity of these compact systems for whatever applications they might be suitable for. So now half the company works on diagnostics, and the other half on therapeutics. We also have a small team working on core discovery and biochemistry of CRISPR systems.
Half the size does the whole job
So from the very beginning, you have focused on very compact Cas nucleases. Cas14 is only one third the size of Cas9 so how can it still do its job?
This is an excellent question, and it puzzled us for a while. When the structure of these compact effectors was published it was demonstrated that they form a homodimer. So two of these Cas14 subunits come together and scaffold under the guide RNA and make a complex that is much more reminiscent of what you might think of as Cas9 or Cas12a.
There have been some efforts to chop away a piece of Cas9 and make it smaller. Remarkably, the homodimer formation is a solution that you would never come upon if you were doing protein engineering. But the natural diversity that has taken hundreds of millions of years of evolution has been able to hone this really elegant solution to the size problem.
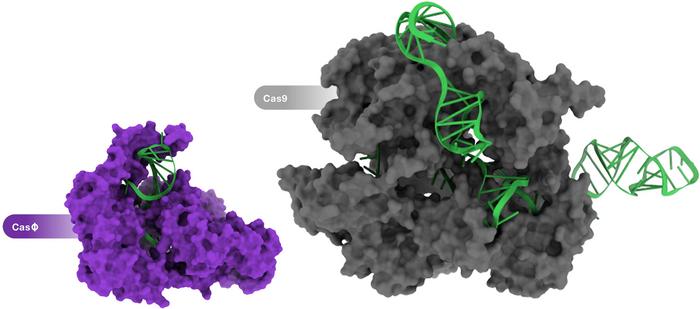
So the homodimer formation is the reason that Cas14 can be less than half the standard size?
Yes, exactly. The functional protein still weighs about the same as a regular CRISPR system, but encoding it in nucleic acids will be much more efficient. All these small CRISPR systems come from weird branches of the tree of life, like nano archaea and other kinds of bacteria that are smaller than some viruses. So there has been tremendous evolutionary pressure to make these systems as efficient as possible in terms of genome size. And so they landed on this solution.
CasΦ discovery and characterisation
CasΦ was first reported last year in an article in Science by researchers in Jennifer Doudna's and Jill Banfield's labs at the University of California, Berkeley. The team discovered CasΦ as a new member of the CRISPR-Cas arsenal that originates from the largest known bacteriophages. Characterisation revealed that CasΦ targets and cleaves bacterial, human and plant DNA and that it is only about half the size of Cas9, which make it a very attractive addition to the gene-editing toolbox. We interviewed joint-first authors Basem Al-Shayeb and Patrick Pausch PhD from Doudna's lab shortly after this work was published. Earlier this year, Patrick Pausch was joint-first author on a publication in Nature Structural and Molecular Biology, which reported the cryo-EM structures of CasΦ (Cas12j) in pre- and post-DNA-binding states. The other first author on this work was Katarzyna Soczek PhD, a postdoctoral fellow in Eva Nogales' lab, also at the Universiy of California, Berkeley.
Fast rejection leads to higher accuracy
Does the small size come with benefits or drawbacks?
The key characteristics we are looking for in any Cas system are efficacy, i.e., actually cutting the DNA efficiently, and safety, i.e., minimal or no off-target activity.
For all of these compact systems, there are still tens of thousands of different ones to choose from - some of them are higher fidelity, and some are lower fidelity - and it is really about mining all of them until you find the right system that has the features you want. We have been able to find much more accurate systems than the gold standard of Cas9 in terms of fidelity.
Is the small size of these compact systems the reason for their higher accuracy?
I think the accuracy has more to do with the mechanism by which the Cas recognises its substrate. Cas9 has this pretty complicated system where different protein domains look at the substrate and see if there is a target match. These small systems are a little bit more straightforward; they quickly reject off-targets as opposed to sitting on them and having a smart, complex mechanism. So if it is not a match, they just kind of spit it back out again.
What are the differences and similarities between the small Cas nucleases?
There is definitely quite a bit of diversity. Within the different platforms we are working on at Mammoth, we try to leverage various features. Some of them have non-specific collateral activity and are suitable for diagnostics. Others only do double-stranded DNA cleavage and some target RNA, etc. In the end, the useability of each platform depends on the disease target and the kind of edit we need to make.
Most of the CRISPR therapies in development are based on Cas9. Do you envision that we will see much more specialised Cas nucleases used for different diseases or different purposes in the future?
For regulatory purposes, you would still want there to be multiple uses for one system. But, still, I think many targets are inaccessible with the first-generation systems that use SpCas9 or Cas12a.
Sometimes you want to use a technique that doesn't make indels, or you want to knock down RNA or make a different type of integration. It isn't one size fits all. A lot of the hype about CRISPR is that it is a general tool that can cure all diseases, but in reality, it will take multiple systems to deliver on that. This is where the CRISPR toolbox approach comes in.
Compact size leaves room for additional cargo
Cas14 and CasΦ are variants, but do you also see diversity within each group of Cas nucleases?
Yes. Even within Cas14, there are thousands or tens of thousands of variants with different features and properties. Same with CasΦ, there is quite a bit of diversity within that also. Some of the diversity is natural, but you can also use protein engineering to create even more diversity. Some companies are very focused on engineering, while others are more focused on natural diversity. At Mammoth, we see them as complementary skill sets. There are specific problems that engineering can solve more quickly than natural mining. And for other issues, the opposite is the case, for instance the homodimer of Cas14, which would never be possible with engineering.
I can't give an exact number, but there are tens of thousands of different natural variants. And that number keeps growing as more and more metagenomic projects explore the microbial dark matter out there.
“The systems we are looking at are very compact, so you are only using half the AAV. This means that the other half is left for some of the more precise methods for gene editing, e.g., base editing or prime editing that take up extra capacity”Lucas Harrington
Can you give any examples of specific Cas variants that might be best for certain purposes?
Well, we haven't disclosed our pipeline publicly, so I can't dig too much into that. We just announced our partnership with Vertex, but we also haven't disclosed this pipeline. But I think AAV is a concrete example of this. With first-generation systems, you can just barely fit the nuclease and the guide into the AAV. Still, the entire capacity is just encompassed by having a double-stranded break and that is just going to do very traditional cutting.
The systems we are looking at are very compact, so you are only using half the AAV. This means that the other half is left for some of the more precise methods for gene editing, e.g., base editing or prime editing that take up extra capacity. It is only with the small Cas systems that we can pack all these precise gene-editing tools into an AAV. Since AAV is the first choice for delivery to the muscles, the heart, the eye and the CNS, you can start leveraging that delivery platform if you choose the right CRISPR system from the catalogue of variants.
Can you give an example of a disease and a Cas variant that go especially well together?
Sure, let’s take diseases in the muscles. The muscles are a critical area where you want to use the right serotype of AAV for systemic delivery and need to pick a CRISPR system that plays nicely together. For example, in the case of Duchenne muscular dystrophy, you might want to use exon skipping, and so you need to make a pretty large deletion and make sure that you are knocking out the junction very efficiently. Some systems make minimal changes, and others make larger changes to the genome, so you choose the variant that suits your purpose best.
Can you disclose anything about what disease you are working with or will be working with?
I can't get into too many details, but generally, we are very focused on working with in vivo therapies because of our enzymes' features. Here we can use our compact systems to get things to the target organs. This is where we see our most significant differentiation and want to make our most prominent indications. Apologies that I can't disclose any specific diseases that we are going after, but that's the general parameters of what we are focused on.
CRISPR can deliver true precision medicine
Can you maybe tell me how far you are in the process and when you expect to disclose something?
That will hopefully be coming not too far into the future, but it depends on many things. We are pre-clinical, obviously, and still establish the systems and testing them for specific indications. Once we are very confident that there is an ailment that we are going after, we will communicate that. So we don't have a hard deadline, but I don't think it will be too far into the future.

Where do you see the most significant potential for CRISPR in the future? Not just for Mammoth but the whole medical field?
I think it's very early days for the CRISPR field in general. We have decades of work ahead of us to deliver on the promises of this platform. I think it is not just going to be a technical advancement. What CRISPR enables is true precision medicine, where you are doing gene editing at a personalised scale.
That's not just a technical challenge but also a regulatory challenge. How do we deal with something where you might have ten different drug products targeting different genotypes of the same disease? Here you will not have to do a complete process for each of those compounds; the regulatory bodies can recognise that you are just changing the guide RNA. You have done some safety assessments on the guide RNA, but the edits they are making are essentially the same. I think that is really where the problem is. It's going to take a long time, and rightfully so, we need to demonstrate the safety of these systems.
That is where I see a promise of CRISPR. Having a system that allows us to treat diseases that aren't just the largest and affect hundreds of thousands of people, but much smaller patient populations.
Do you think it would be practical to tailor treatments to specific patients? Wouldn't that come with a high price tag?
I think that's where things are going with CRISPR. If you think of older gene-editing techniques like zinc fingers and TALENs, personalised treatments are not practical because each edit requires a different nuclease. With CRISPR, we can spin out new therapeutic candidates very quickly because the system's power is that it is highly programmable.
However, there will need to be a different way of looking at such treatments from a safety perspective. If you have a patient population of a hundred patients, it becomes challenging to run a clinical trial and actually demonstrate safety and efficacy. Even with cystic fibrosis, a single mutation causes the disease in most patients, but rareer mutations also can benefit from gene editing and in an ideal world we can help all patients, not just a subpopulation. We need to be able to treat those more rare versions of the disease, and we have the potential to do so with CRISPR.
The future lies in in vivo applications
You mentioned before that Mammoth is very focused on in vivo systems for practical reasons. Do you think this is also where the future lies?
Yes. It's not that we are against ex vivo therapies by any means, but I think in vivo is just where things need to go, if we can do it safely. The challenges of doing ex vivo therapies are taking cells out, editing them, expanding them, conditioning the patient, putting the cells back in. With in vivo therapies, you might only need a single intravenous injection. That would really be transformative in terms of how widespread these treatments could become.
“I think it's achievable. It can be done”Lucas Harrington
Even very established approaches such as CAR-T therapy, if you can move that in vivo, imagine the number of patients you could access, how much kinder the treatment would be to the patients, and how much quicker you could do it. I think this is a tremendous opportunity
But with in vivo therapies, you need to be very, very sure of what is happening inside the patients. So how will you handle that?
If you're delivering something systemically and targeting every cell in the body, the risk of off-targets is an immediate concern. But you can reduce that risk if you are targeting a single organ and only edit the exact cells that you are looking at. I think a lot of that is about delivery again.
So with systemic delivery, you need to think very differently about the safety versus ex vivo where you take a particular type of cell out, edit it and put it back in again - it's a much more controlled setting. But I think it's achievable. It can be done.
Link to relevant publications on small Cas nucleases:
New CRISPR–Cas systems from uncultivated microbes
Programmed DNA destruction by miniature CRISPR-Cas14 enzymes
Editorial note: This interview article was first published on the 22th November 2021. A fact box entitled: 'CasΦ delivery and characterisation' was added on the 24th November 2021 to reflect important contributions by researchers at University of California, Berkeley to our current understanding of CasΦ.
Tags
ArticleInterviewNewsAdeno-associated virus (AAV)Cystic FibrosisDuchenne Muscular Dystrophy, DMDCas14Cas9CasΦMammoth Biosciences, Inc.
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







