Multiplex CRISPR Merges With Click Chemistry
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
Do you plan to organise a webinar?
Why not use the CMN platform and get exposed to the global gene editing community.
More details here.

»When I started my research group, we were disappointed to find out that you couldn't just go to your favourite vendor right now and say, I would like a guide RNA for gene X, Y, and Z. And I would like them with three different fluorophores so that I can do a three-dimensional CRISPR assay,« says Matthew Macauley, who is an associate professor in the Department of Chemistry at the University of Alberta, Canada.
He refers to multiplex CRISPR experiments where you want to select for cells that have taken up all three single guide RNAs (sgRNAs) and thereby increase the probability of isolating triple-edited cells. He teamed up with his colleague in the same department, associate professor Julianne Gibbs, who came up with an innovative synthetic solution for developing a fast and straightforward method for synthesising complex sgRNAs.
The method uses a modular approach, synthesising each sgRNA in three separate fragments of 41 nucleotides (nts) or less. Each piece is synthesised with special reactive groups at the 5'- and 3'-prime ends that make them snap together in a process called click chemistry. The modular approach allows for shuffling parts containing the targeting region, a fluorescent marker, and other modifications. Thereby, various sgRNAs with different functional characteristics can easily be assembled. Julianne Gibbs and Matthew Macauley are co-senior authors of a paper published earlier this year in Bioconjugate Biochemistry. They believe their strategy will be well suited for custom, scalable synthesis of sgRNAs that can be used in multiplex CRISPR experiments.
Click Chemistry
Click chemistry refers to a group of fast, simple, and high-yielding biocompatible reactions that join small modular chemical units or, e.g., biomolecules and reporter molecules. The “click” can be seen as the two pieces of a seat belt buckle that easily snap together. Each piece is attached to either of the two molecules to be joined, and upon mixing, they conjugate with high specificity and minimal by-product formation. Several click reactions can be used, but a popular choice is an azide and an alkyne that quickly and irreversibly form a triazole linkage. One advantage of this system is that azides and alkynes rarely occur in biological systems, thereby minimising the risk of unspecific reactions.
Correct clicking is ensured by DNA-RNA hybridisation
A traditional sgRNA is a fusion of two naturally occurring RNAs of the bacterial CRISPR immune system: a virus-derived CRISPR RNA (crRNA) that contains a 20-nt programmable targeting sequence; and a bacterially-encoded trans-activating crRNA (tracrRNA) that binds Cas9. If synthesised in one piece, the solid-phase synthesis efficiency of a typical sgRNA of 97 nts will only be around 5%. That efficiency is reduced even more if fluorescent modifications are introduced. This can lead to such small quantities of the sgRNA material that it cannot be thoroughly purified or characterised by gel electrophoresis and mass spectrometry.
“The middle piece is where we insert the fluorophore, so if we want to edit three genes simultaneously, we make three middle pieces with different fluorophores and then swap those in and out. Then we swap the 5´-prime ends containing different targeting sequences in and out, and there you go. That is the real advantage of our method”Matthew Macauley
»We think that as CRISPR sees greater use, people will want to combine the guide materials to target many targets at once, and then you’ll need larger amounts of material. And one of the challenges of making the guide RNAs by solid phase synthesis is that they have a length where you’re starting to take really big hits in the amount of material you can make,« explains Julianne Gibbs.
To solve that problem, the two researchers and their colleagues looked for two suitable sites to divide the sgRNA into three short fragments that can be synthesised easily. And subsequently, the three fragments should be stitched together by the click chemistry system without affecting functionality. After a series of initial testing, they settled on three fragments of lengths 30, 41, and 27 nt. This divided the sgRNA at C30:T31 in the crRNA-region near the gene targeting sequence and C70:T71 around the middle of the tracrRNA region (Figure 1B).
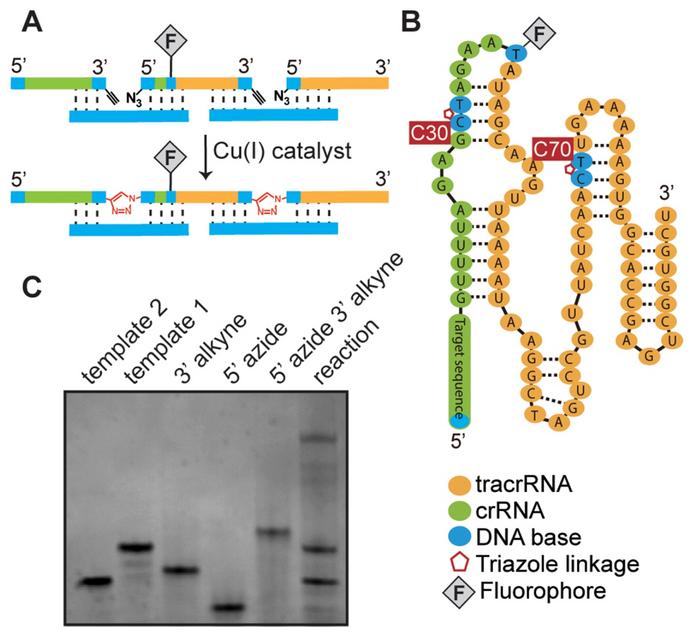
The fragments were synthesised and terminated with clickable 3’-O-propargyl- and 5’-azido-modified deoxyribonucleotides. These are alkynes and azides, respectively, and will eventually snap together to form a triazole linkage. In addition, an extra dT nucleotide was inserted in the tetraloop region of the middle fragment between A35 and A36, allowing for other applications such as linkage to a fluorophore.
All three strands were clicked simultaneously in the presence of a copper catalyst using two DNA templates that brought the desired azide and alkyne termini in proximity and prevented unwanted by-products (Figure 1A & 1C). The DNA templates assured correct assembly while control experiments without templates generated less product and more by-products.
Double-clicked sgRNAs allow for flexible multiplex CRISPR
Subsequent in vitro DNA cleavage assays using a double-clicked sgRNA targeting the HRPT gene showed that the sgRNA was well tolerated by Cas9 and led to cleavage activities comparable to an unmodified sgRNA. This confirmed that the two triazole linkages at C30:T31 (close to the targeting region) and C70:T71 (close to the Cas9 binding site) did not interfere with targeting or cutting.
Cas9-expressing 293T cells were transfected with the double-clicked sgRNA using lipofectamine to test activity in living cells. The T7 endonuclease I assay showed that the double-clicked sgRNA exhibited better cleavage (13%) than in vitro transcribed sgRNA (6%). However, commercial sgRNA showed higher cleavage (27%), probably due to proprietary modifications stabilising the guides in the cellular environment.
Further experiments demonstrated the flexibility of the modular sgRNAs that allow for multiplex CRISPR experiments. The researchers synthesised two versions of the 5'-prime fragment containing the targeting region, so they targeted the EMX1 and WAS genes, respectively. The middle fragment containing the functionalised extra dT between A35 and A36 was also synthesised in two versions with either an ATTO 550 or a fluorescein fluorophore attached. These fragments and the 3' fragment were combined by click chemistry to form ATTO 550-labelled sgRNA targeting EMX1 and fluorescein-labelled sgRNA targeting WAS (Figure 2A).

The two double-clicked sgRNAs were transfected into Cas9-expressing 293T cells, either individually or together (Figure 2B). Transfected cells were then sorted by fluorescence-activated cell sorting (FACS) according to which of the two fluorophores they had taken up. These experiments showed that most of the cells took up each of the sgRNAs when transfected individually, while around 40% of doubly-transfected cells took up both sgRNAs (Figure 2C).
Combining synthetic sgRNAs with different fluorescent markers and FACS cell sorting is useful in multiplex CRISPR gene editing since it allows easy selection of cells that have taken up all the sgRNAs. This enhances the probability of isolating cells containing all the desired edits. And the study by Julianne Gibbs and Matthew Macauley demonstrates that click chemistry can contribute to multiplex gene editing by allowing for a fast, efficient, and modular strategy for synthesising a variety of sgRNAs with different targeting regions and fluorescent markers.
»The middle piece is where we insert the fluorophore, so if we want to edit three genes simultaneously, we make three middle pieces with different fluorophores and then swap those in and out. Then we swap the 5´-prime ends containing different targeting sequences in and out, and there you go. That is the real advantage of our method,« concludes Matthew Macauley.
Link to the original article in Bioconjugate Chemistry:
CRISPR-Click Enables Dual-Gene Editing with Modular Synthetic sgRNAs
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsDeliveryIn vivoAdeno-associated virus (AAV)Adenovirus (AV)DiseasePhenylketonuria (PKU)Prime editors
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







