Safer, More Efficient AAV Systems for Therapeutic CRISPR Genome Editing

Adeno-associated viral vectors (AAVs) are currently the most widely used delivery vehicles for in vivo genome editing, however, their therapeutic application is significantly restricted by cargo limits. AAVs are small – around 4.7kb – making it difficult to package all the necessary gene-editing components into a single vector.
There have been many attempts to address the issue of AAV cargo limits. One of them is to employ smaller Cas9 variants that have been discovered in recent years, such as Cas9 from Staphylococcus aureus (SaCas9), but these compact CRISPR systems come at the cost of reduced editing efficiencies and a smaller range of potential target sequences owing to their PAM requirements.
In a study published last month in Nature Communications, members of Professor Erik Sontheimer’s lab at the University of Massachusetts Chan Medical School demonstrated the use of a cleverly engineered, all-in-one, self-inactivating AAV vector with a compact Cas9 variant for CRISPR therapeutic genome editing in vivo.
First author Dr. Raed Ibraheim joined the Sontheimer lab in 2015 as a PhD student with a passion for AAV vectors after spending a couple of years producing clinical-grade AAVs in a GMP manufacturing facility. Ibraheim’s AAV:Nme2Cas9 system enables therapeutic genetic modifications mediated by both non-homologous end-joining (NHEJ) and homology-directed repair (HDR), and can therefore be used to treat multiple genetic diseases.
Commenting on the significance of this work, Ibraheim says: »We’ve shown that you can make these self-inactivating AAVs. You can clone, test, and package them without problems, and they can provide therapeutic benefit in these proof-of principle disease models. Our goal was to make AAV:Cas9 vectors that, once they’ve done their job, will self-eliminate and disappear. I hope there will be more ways to build on this technology in the future - there’s a huge benefit if we spend more time making AAVs more safe and more active.«
Nme2Cas9: A triple-threat Cas nuclease
The Sontheimer Lab were early pioneers of the use of a Neisseria meningitidis Cas9 variant (Nme1Cas9) for genome editing in vivo in induced pluripotent stem cells (iPSCs). While Nme1Cas9 is smaller than the commonly used Streptococcus pyogenes Cas9 (SpyCas9) and is therefore more amenable for packaging into AAVs, its required protospacer adjacent motif (PAM) sequence is longer, meaning its range of genomic targets will be reduced.
This problem was the first of several that the team aimed to overcome; they needed a Cas nuclease with the short PAM requirement and high editing efficiency of SpyCas9, but with the compact size of Staphylococcus aureus Cas9 (SauCas9) or Nme1Cas9. As it happens, they eventually found it in a different strain of the same N. meningitidis bacteria – Nme2Cas9 – and this work was published in a 2019 paper in the journal Molecular Cell.
The appeal of Nme2Cas9 is three-fold: it’s small, making it easier to package into AAVs; it has a dinucleotide PAM sequence, meaning it has a broader range of genomic targets; and it displays high editing efficiencies in mammalian cells with low off-target activity, making it ideal for therapeutic applications. Ibraheim and his colleagues knew they were on to a winner.
»With Nme2Cas9, it’s also small, it also fits into the AAV capsid, but it’s highly accurate – it doesn’t have the propensity to cut at off-target sites like wild-type SpyCas9. The other advantage is that the PAM is small. It’s a dinucleotide PAM just like SpyCas9,« Ibraheim elaborates.
Overcoming packaging limits for AAVs, one step at a time
Most studies that use AAVs to deliver gene therapy attempt to fit one Cas9 nuclease and one single guide RNA (sgRNA), along with their respective promoters, into the vector in order to create a single double-stranded break in DNA. Given the 4.7kb packaging limit of most AAVs, this is usually a significant challenge, particularly for studies that aim to correct genetic mutations by providing a donor DNA template for HDR.
Many studies have addressed this issue by employing a multi-vector strategy, dividing the CRISPR components between two AAV vectors and delivering them simultaneously to achieve editing. It seems simple enough, however there are significant safety concerns regarding the toxicity of high AAV dosages, let alone the combined AAV dosages administered in multi-vector strategies.
Ibraheim’s study takes this packaging problem one step further, aiming to create a vector with sufficient cargo space to fit two sgRNAs rather than just one (Figure 1). By doing so, CRISPR could be used to excise pathogenic trinucleotide repeat expansions, like the one that causes Friedreich’s Ataxia.
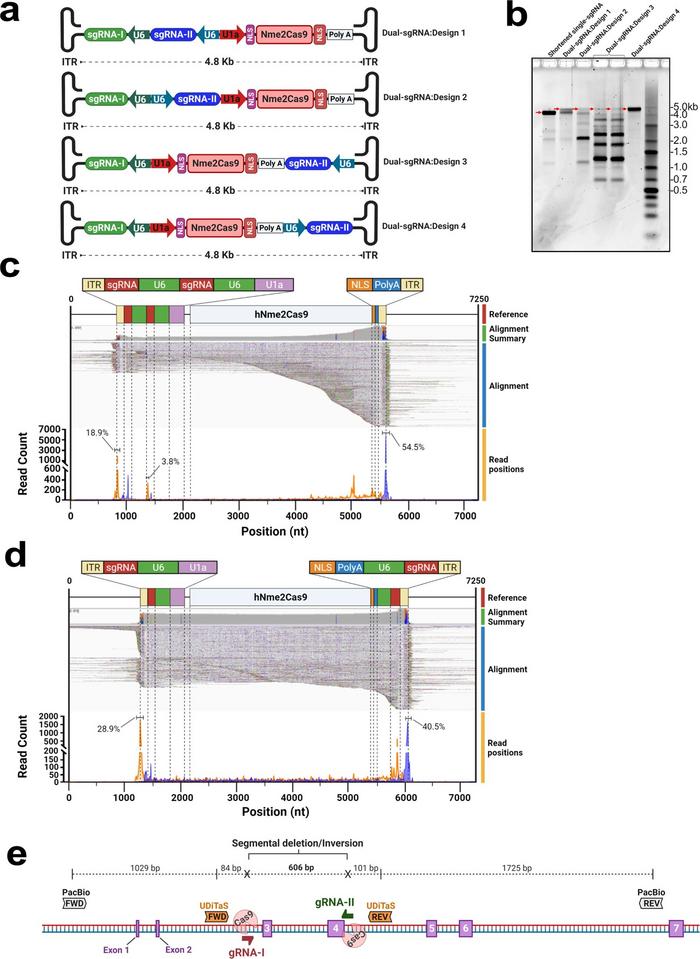
Alternatively, they could also deliver a single sgRNA along with a donor DNA template for HDR to correct pathogenic mutations, negating safety concerns regarding the multi-vector strategy. To free up space in the AAV vector, the team took a step-by-step approach.
»We started to go through the different components of the first version of this AAV-Nme2Cas9 that we had created in our study from 2019. We engineered the guides, and we were able to truncate them from 145 down to 100-120 nucleotides. We also modified the other elements, piece by piece. For example, before, we had four nuclear localisation signals, but we wondered if we could keep only two of them while maintaining efficient Nme2Cas9 activity,« Ibraheim explains.
By addressing each individual component of the system, Ibraheim and his colleagues were able to create an AAV that is about 4.4kb, including an sgRNA, Nme2Cas9, and promoters. They were left with a ~400 nucleotide space, into which they could insert either a second sgRNA or a donor DNA template. It was a big win for the team, but Ibraheim was only just getting started.
Anti-CRISPR proteins for self-inactivating AAVs
The next step of the project was to address a long-standing problem in gene therapy. If editing components are expressed long-term within cells, they can pose risks to patient safety, including immunogenicity and off-target editing.
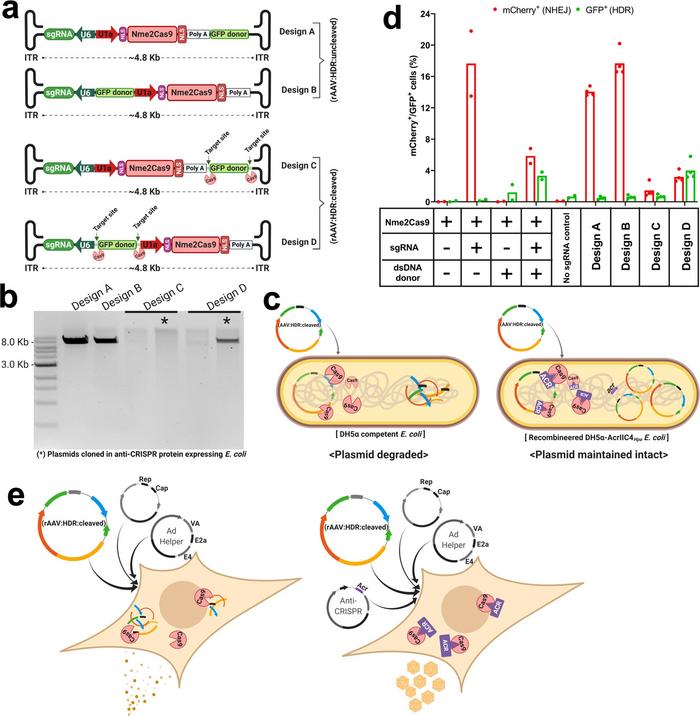
»When AAVs are delivered inside cells, they form episomal concatemers in the nucleus and this can lead to long-term expression of the Cas9, while in most cases, a transient expression of Cas9 [for therapeutic editing] is sufficient. Our goal was to engineer an all-in-one AAV:Cas9 vector that has the ability to cut the target site, correct disease-causing mutations by HDR, and subsequently inactivates itself by the function of its own Cas9,« Ibraheim describes.
The team added Cas9 target sites flanking the HDR donor DNA template, with the hope that Cas9 would then cut the donor from the AAV and make it linearised double-stranded DNA, which would help the HDR process and would also enable self-inactivation of the vector. But at this stage, Ibraheim ran into an obstacle.
»While I was trying to clone these self-inactivating AAV:Cas9:HDR plasmids in bacteria, we faced a major hurdle. When we added the target sites flanking the HDR donor, we couldn’t make the cloning work. All the plasmid was being degraded, so I didn’t see any traces of it on an agarose gel. We eventually realised that there was leaky expression of Cas9 in the E. coli competent cells that we were using for cloning, and Cas9 was cutting the plasmid itself,« Ibraheim comments.
The team did some troubleshooting, and eventually decided to tackle this issue by employing anti-CRISPR (Acr) proteins. Acr proteins are produced by viruses and mobile genetic elements and impede CRISPR-Cas systems. Ibraheim co-transformed the AAV plasmids containing the extra target sites with another plasmid coding for Acr proteins, but there was one more obstacle.
»These [Acr] proteins bind to the Cas9 and inactivate it so it can’t cut. It worked – we could now see the plasmid on the gel if we co-transformed an Acr-expressing plasmid in trans. The issue was, if we want to package a functional self-inactivating AAV:Cas9 plasmid, we had to find a way to make the cloning competing cells endogenously express Acr protein. So we decided to engineer our E. coli competent cells,« Ibraheim says.
Ibraheim’s UMass colleague, PhD student Samantha Nelson, used λred phage recombineering technology to insert a cassette that codes for an anti-CRISPR protein into the chromosome of E. coli (Figure 2). This prevented the AAV:Nme2Cas9 vector from self-cutting so that it would stay intact during the cloning process, but could still self-inactivate when delivered in vivo. Acr proteins were also used during the packaging of those plasmids in AAV8 and AAV9 capsids to prevent the self-cutting of those plasmids during AAV manufacturing.
Nanopore sequencing pipeline for profiling AAV vectors
The study had another notable output in the development of a novel nanopore-based sequencing pipeline designed to profile AAV genomes in order to maximise safety and efficiency for clinical translation. As Ibraheim explains, the packaging of AAV vectors is not straightforward, particularly when attempting to package multiple sgRNAs:
»Working closely with our collaborator Dr. Phillip W.L. Tai, we found that since the sgRNAs have short hairpin structures, having more than one guide can lead to the formation of strong secondary structures inside the vector. Depending on the location and orientation of these guides, the packaging efficiency of the vector will change. We ended up with some arrangements where some of the capsids had a truncated genome with only the Cas9, and no guide. Others had the guide and only half of the Cas9. Obviously, this is bad news because when packaging efficiency is reduced, it will have an impact on vector integrity and the safety of these therapies,« Ibraheim says.
The team collaborated with Professor Guangping Gao’s lab, also at UMass, to develop a nanopore-based sequencing pipeline for AAVs that would help determine the best position and orientation to place these guides when engineering dual-guide RNA AAV:Cas9 vectors. Ibraheim also hoped to use this sequencing platform to improve quality control measures for AAVs and increase patient safety.
»If we can make pure and active AAV particles, then we don’t need to deliver a high dose. This will also help to reduce the immunogenicity and the high cost that is associated with developing and manufacturing these vectors,« Ibraheim asserts.
Testing self-inactivating AAV:Nme2Cas9 vectors in vivo
To demonstrate the AAV:Nme2Cas9 vectors would work in vivo, Ibraheim and his colleagues tested the system in two different murine disease models. The team used the AAV:Nme2Cas9 vector containing two sgRNAs to efficiently knock out the Hpd gene in the tyrosine degradation pathway and rescue the lethal phenotypes in a mouse model of Hereditary Tyrosinemia type I (HT-I).
The team then used the AAV:Nme2Cas9 vector with one sgRNA and a donor DNA template to correct two pathogenic point mutations: Fah in a second mouse model of tyrosinemia, and Idua in a mouse model of Mucopolysaccharidosis type I (MPS-I) via HDR. In both, editing was successful and editing efficiencies were therapeutically relevant.
Ibraheim also checked to see if the vector would indeed self-inactivate in vivo, and he was very happy with the results.
»The number of AAV particles inside the mice that received the self-inactivating AAV is lower compared to those that received the normal AAV. We also show there is a much lower amount of NmeCas9 and guide RNA transcripts,« Ibraheim emphasises.
Ibraheim defended his thesis in November of 2020 and now works at Tessera Therapeutics in Somerville, MA. He continues to work on various delivery modalities of gene therapy and remains hopeful that the field of AAV gene delivery will continue to grow by focusing on patient safety.
Ibraheim also offers some sage advice to new graduate students, highlighting the importance of teamwork and a supportive lab environment when struggling with problems in the lab.
»In my opinion, only half of a PhD training is about learning the science. The other half is all about us, the PhD students. Attributes, such as learning to push through the difficulties, growing as creative thinkers, and training ourselves to be proactive and collaborative, are some outcomes of a successful PhD training. Having a supportive advisor and colleagues in the lab were really valuable in shaping my own PhD experience. I highly encourage new graduate students to spend more time talking to other researchers. You don’t have to solve every problem alone - teamwork is important.«
Link to the original paper published in Nature Communications: Self-inactivating, all-in-one AAV vectors for precision Cas9 genome editing via homology-directed repair in vivo.
Rebecca Roberts is a molecular biologist and science writer/communicator based in Queensland, Australia.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsin vivoIn vivoAdeno-associated virus (AAV)Mucopolysaccharidosis I, MPS IRare DiseaseCRISPR-CasCas9
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







