Using CRISPR-Cas9 to “Attack” Cancer Cells: A New Avenue for Personalised Cancer Treatment?

Despite recent advances in cancer treatment using drugs that induce DNA damage or boost anti-tumour immune responses, eliminating tumour cells whilst leaving normal cells intact remains a major challenge.
In a recent proof-of-concept study, researchers from the Institute of Basic Science and Ulsan National Institute of Science and Technology (UNIST), Republic of Korea, developed CINDELA, a new CRISPR-Cas9-based method for selectively inducing DNA damage and cell death in cancer cells.
»We developed an innovative CRISPR-Cas9-based approach to target multiple cancer-specific mutations that are accumulated in cancer cells during tumour development,« said Taejoon Kwon, PhD, Associate Professor at UNIST and first author of the study. »Using this method, we can introduce multiple DNA double-strand breaks (DSBs) and induce cell death selectively in cancer cells,« he added. The team’s findings were published in PNAS late last month.
Dr Kwon also noted that they validated the ability of CINDELA to selectively eliminate tumour cells in cancer cell lines, patient-derived cancer cells, mouse models, and patient-derived xenografts. This extensive validation makes him confident that the novel approach may open new avenues for personalised cancer treatment.
Dr Sandra Rodriguez-Perales, an expert in the use of CRISPR-Cas9 to engineer tumour cells, agrees that there is a clear need for new therapeutic alternatives that can selectively target cancer cells without affecting healthy cells. She is the head of the Molecular Cytogenetics Unit, Spanish National Cancer Centre (CNIO) and did not participate in the study.
»This study has advanced a little further along the long road that could lead to the future treatment of different types of tumours,« she said.

Role of genomic instability in cancer
Most cancer cells exhibit a high degree of genomic instability, which leads to the accumulation of alterations in genes and regulatory regions of the genome in cancer cells. These alterations range from single-nucleotide mutations to large-scale structural rearrangements in chromosomes. Small insertions and deletions (InDels) are genomic alterations that are frequently found in cancer cells but not in neighbouring non-malignant cells.
Genomic instability is therefore characterised as a hallmark of cancer, and Dr Kwon pointed out that this plays a key role in cancer cell evolution and tumour progression: »Most tumour cells accumulate thousands of InDels, providing an opportunity to selectively eliminate cancer cells by targeting InDels that are present in cancer cells but not in normal cells.«
Importance of cancer cell selectivity in cancer treatment
In contrast to infectious agents that are foreign to the human body, tumours develop from our own cells; this makes it difficult for anti-cancer agents to distinguish cancer cells from normal cells.
Conventional cancer treatments, including chemotherapy and radiation therapy, kill rapidly proliferating cancer cells by inducing DNA damage and inhibiting DNA replication. Despite their widespread use, their drawbacks are well recognised: »These conventional treatments do not discriminate cancer cells from non-malignant cells; hence they induce DNA damage and cell death in neighbouring normal cells, causing many side effects,« Dr Kwon explained.
Identifying features that are specific to cancer cells is essential for the development of new treatments to effectively eliminate cancer cells whilst sparing healthy cells. The team hypothesised, therefore, that InDels accumulated in cancer cells might represent an actionable feature specific to cancer cells.
Repurposing CRISPR-Cas9 to develop a cancer-specific InDel “attacker”
Guided by CRISPR RNA guides, Cas9 endonuclease introduces DNA double-stranded breaks (DSBs) into specific DNA sequences. Exploiting this principle, Dr Kwon and his colleagues developed a CRISPR-Cas9-based method to selectively eliminate cancer cells by targeting cancer-specific InDels and introducing multiple DSBs specifically in cancer cells. They termed this method “cancer-specific InDel attacker", or simply “CINDELA” (Figure 1).
DSBs are very harmful to cells and are often considered an undesirable effect of CRISPR-Cas9 editing. However, the team realised that this “side effect” of CRISPR-Cas9 may, in fact, be useful in the selective targeting of cancer cells.
“Although the concept of CINDELA sounds very obvious, this is the first proof-of-concept study to show that it is feasible to use CRISPR-Cas9 to selectively kill cancer cells by targeting cancer-specific InDels”Taejoon Kwon
»By targeting cancer-specific InDels, we can introduce multiple DSBs in the genome, which, if not repaired, can lead to cell death,« Dr Kwon said. He also explained that, compared to single-nucleotide mutations, InDel mutations are less prone to the off-target effects of CRISPR-Cas9.
»Although the concept of CINDELA sounds very obvious, this is the first proof-of-concept study to show that it is feasible to use CRISPR-Cas9 to selectively kill cancer cells by targeting cancer-specific InDels,« Dr Kwon added.
Using CRISPR-Cas9 to induce multiple DSBs induces cell death in cancer cells
As a proof-of-concept, the researchers used CRISPR-Cas9 to simultaneously introduce multiple DSBs into the genome of cancer cells. They achieved this by targeting well-known CRISPR-Cas9 target sequences that appear repeatedly in the human genome.
They used lentiviruses to deliver increasing numbers of multiple-target sgRNAs and Cas9 into various types of cancer cell lines, including HCT-116 (colorectal carcinoma), MDA-MB-231 (breast adenocarcinoma), and K562 (chronic myelogenous leukaemia). Expression of multiple-target sgRNAs in all these cancer cell lines led to a significant reduction in cell survival, with cell fitness ranging from 80% to nearly 0%. The extent of cell death was greater when a higher number of sgRNAs was used.
Targeting cancer-specific InDels induces DSBs and apoptosis in cancer cell lines and patient-derived cancer cells
To identify cancer-specific InDels, Dr Kwon and colleagues analysed the whole-genome sequences of U2OS (osteosarcoma) and HCT-116 (colorectal carcinoma) cells, as well as a non-tumour epithelial cell line (RPE1). Comparison of these sequences led to the identification of InDels that were unique to each of the cancer cell lines analysed.
They then designed sgRNAs targeting 30 selected InDels that were specific to U2OS cells. Transfer of CRISPR-Cas9 ribonucleoprotein (RNP) complexes that targeted U2OS-specific InDels triggered cell death in U2OS cells but not in HCT-116 cells. No death was observed in U2OS cells when cells were transfected with CRISPR-Cas9 RNP complexes targeting HCT-116-specific InDels, confirming that selective cell death could be achieved by targeting cell line-specific InDels.
To overcome challenges associated with the in vivo delivery of RNP complexes to the tumour microenvironment, Dr Kwon and colleagues used adeno-associated virus serotype 2 (AAV2) carrying Staphylococcus aureus Cas9 (SaCas9) and sgRNAs targeting 30 cancer-specific InDels. Using this AAV-based CINDELA approach, they confirmed that targeting U2OS-specific InDels could reduce cell survival by nearly 80%.
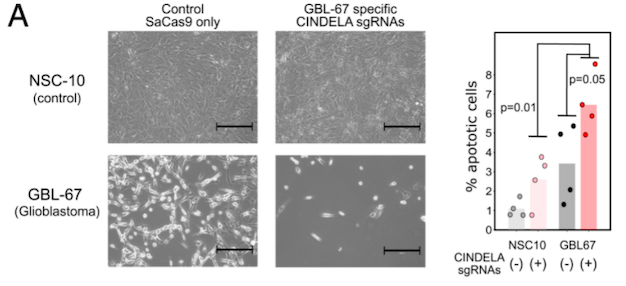
To confirm the feasibility of using CRISPR-Cas9 to selectively kill cancer cells obtained from primary tumours of patients, the researchers performed whole-genome sequencing analysis of glioblastoma cells derived from the primary tumour of a Korean patient and identified 26 cancer-specific InDels. Targeting these 26 InDels using the AAV-based CINDELA approach resulted in the generation of DSBs and cell death in patient-derived glioblastoma cells but not in neural stem cells, which were used as controls (Figure 2).
CINDELA treatment suppresses tumour growth in mice
To test the ability of CINDELA to inhibit tumour growth in vivo, the researchers injected mice bearing HCT-116 tumours with lentiviruses expressing SpCas9 and sgRNA targeting 23 HCT-116-specific InDels. Treatments were carried out daily for two weeks. CINDELA treatment significantly inhibited tumour growth (relative tumour volume at 14 days ~3.7 a.u. for 23 HCT-116-specific gRNAs versus ~5.3 a.u. for two target loci) without causing side effects in mice. These findings suggest that CINDELA selectively inhibited of cancer cell proliferation without causing damage to healthy cells.
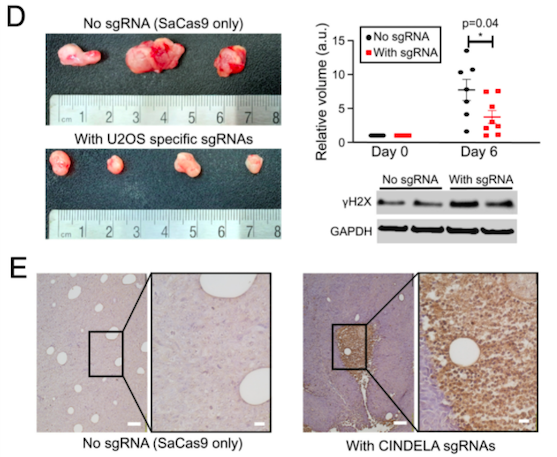
To strengthen their findings, the team treated mice bearing U2OS tumours with AAVs expressing SaCas9 and sgRNAs targeting U2OS-specific InDels. Consistent with their findings in the HCT-116 mouse model, CINDELA treatment significantly suppressed tumour growth compared to control treatment (Figure 3). Additionally, tumours from CINDELA-treated mice showed high levels of the DSB marker γ-H2AX and contained high levels of apoptotic cells.
Finally, the team used AAV-based CINDELA to treat mice xenografted with patient-derived lung tumours after sequencing patient-derived tissues and identifying approximately 30 cancer-specific InDels. Seven days after treatment, patient-derived xenografts in CINDELA-treated mice were significantly smaller than those in the control mice (p=0.033).
Hurdles to clinical development
Dr Kwon noted that strategies to effectively deliver CRISPR-Cas9 components to the tumour microenvironment and regulatory challenges associated AAV dosing are hurdles that remain to be addressed before they can test the ability of CINDELA to eradicate tumours in patients.
Dr Raul Torres-Ruiz, an expert in the development of novel gene editing-based cancer treatments, agrees that in vivo delivery is the most significant hurdle to the widespread clinical success of CRISPR-Cas9 therapeutics. He is a researcher at CNIO and did not participate in the study.
»A high titer of the virus needs to be injected into mice to obtain modest therapeutical results. The low efficiency of in vivo delivery must be enhanced before its therapeutic potential can be fully realised,« he said.
Tumours are heterogeneous and can evolve over time by accumulating new mutations. Consequently, not all cancer cells in a tumour necessarily have the same InDels. Targeting the right InDel mutations is key for the ability of CINDELA to kill all cancer cells in a heterogeneous tumour.
Dr Kwon believes that any surviving cancer cells after CINDELA treatment can be eliminated in a second round of treatment by resequencing tumours and targeting those InDels that survived the first CINDELA treatment.
Looking ahead: Scope for developing combination cancer-selective therapies?
Dr Kwon said that the team is already investigating the mechanisms by which CINDELA induces DNA damage, and they are trying to understand why some cancer cells die and others do not. »There is more heterogeneity in cell death than what we expected,« Dr Kwon explained.
As CINDELA causes cell death following DSB formation, a combination of CINDELA with DNA repair inhibitors may have synergistic effects in inducing apoptosis selectively in cancer cells. PARP1 and ATM inhibitors are examples of agents that can be used in combination with CINDELA to enhance the ability of CRISPR-Cas9 to introduce DNA DSBs. Dr Kwon also mentioned that the team is planning to perform further in vivo validation of CINDELA in patient-derived tumours and in more cancer types.
Link to the original article in PNAS:
Precision targeting tumor cells using cancer-specific InDel mutations with CRISPR-Cas9
Christos Evangelou, Ph.D., is a freelance medical writer and science communications consultant.
To get more of the CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleInterviewNewsIn vivoNon-viralRibonucleoprotein (RNP)ViralAdeno-associated virus (AAV)Lentivirus (LV)CancerSolid TumoursCRISPR-CasCas9
CLINICAL TRIALS
Sponsors:
Suzhou Maximum Bio-tech Co., Ltd.
Sponsors:
Zhejiang University







