Tumour-Specific CRISPR Restores Chemotherapy Response
CMN Intelligence - The World’s Most Comprehensive Intelligence Platform for CRISPR-Genomic Medicine and Gene-Editing Clinical Development
Providing market intelligence, data infrastructure, analytics, and reporting services for the global gene-editing sector. Read more...
NRF2, a master regulator of cellular stress responses, typically helps cells survive oxidative and toxic insults by activating detoxification genes. Mutations that stabilise the protein can turn it into a driver of drug resistance, and this occurs frequently in many tumours, such as lung squamous cell carcinoma.
“We demonstrate that 20%–40% gene editing activity is sufficient to improve response to chemotherapy in animal models”Banas et al.
Kelly Banas, Eric Kmiec and their team focused on a particularly prevalent NRF2 exon-2 mutation (R34G) occurring in the Neh2 domain of NRF2. The mutation disrupts the protein's interaction with its negative regulator KEAP1, leading to protein accumulation and activation of downstream pathways. Crucially, this specific mutation also creates a unique protospacer adjacent motif (PAM) that the CRISPR-Cas9 system can recognise, offering an opportunity for tumour-specific targeting.
Since no suitable cell models existed, they first engineered the R34G mutation into the clinically relevant NCI-H1703 lung cancer cell line using a two-step CRISPR-Cas9 approach that maximised efficiency whilst minimising unwanted modifications (see Figure 1). This generated both homozygous and heterozygous mutant cell lines that served as models for subsequent experiments.
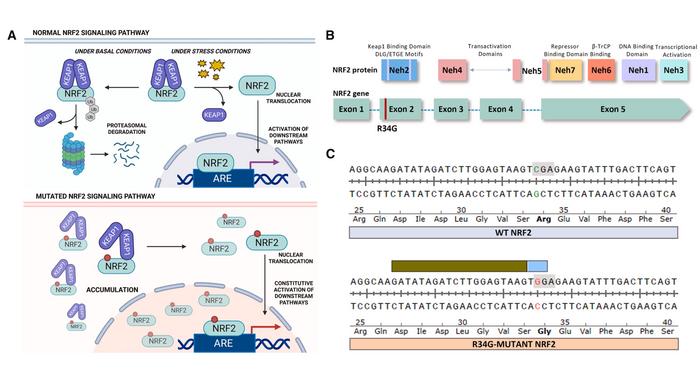
When these engineered cells were treated with CRISPR-Cas9 complexes designed to recognise the R34G mutation, editing efficiencies reached 91.2% in homozygous cells and 38.0% in heterozygous cells. Crucially, when the same CRISPR components were applied to cells carrying wild-type NRF2, virtually no editing occurred, confirming the mutation-specific nature of the approach.
A detailed analysis revealed that CRISPR-Cas9 cleavage produced a characteristic pattern of insertions and deletions, with the most common outcomes being a single-base insertion, a nine-base deletion, and a four-base deletion. The team isolated individual clonal cell lines harbouring different combinations of these modifications to understand how each affected NRF2 function.
Cells containing frameshift mutations that introduced premature stop codons showed complete loss of NRF2 protein, whilst those with in-frame deletions retained some protein expression. Notably, downstream target genes, including NQO1, HMOX1, and GCLC, were significantly downregulated in NRF2-deficient clones, confirming the transcription factor's functional inactivation.
The researchers conducted comprehensive off-target analysis using three complementary methods: in silico prediction, biochemical interrogation, and cell-based assessment. Across 499 nominated potential off-target sites, only five showed detectable editing activity above 0.2%, and this activity was further reduced when using high-fidelity SpCas9 variants.
»We suggest that understanding the genetic diversity of CRISPR outcomes must be a key consideration in identifying effective CRISPR molecules for clinical application,« the authors note in their Molecular Therapy Oncology paper, emphasising the importance of thorough characterisation.
For in vivo delivery, the team selected lipid nanoparticles encapsulating CRISPR-Cas9 mRNA and guide RNA. They screened six different lipid formulations and selected the most promising candidate based on expression levels and biodistribution patterns. When injected directly into tumours in mouse models, the CRISPR nanoparticles achieved editing efficiencies of approximately 20-40%, maintaining the same indel profile observed in vitro. Gene expression analysis from treated tumours showed decreased levels of NRF2, NQO1, and the proliferation marker Ki-67, indicating functional impact on tumour biology.
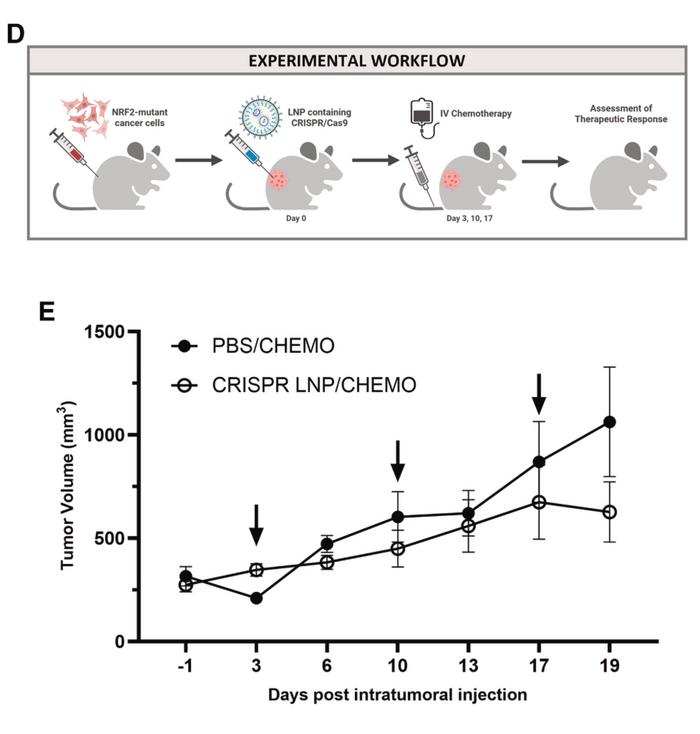
The therapeutic potential became evident in efficacy studies in which tumour-bearing mice received a single intratumoral injection of CRISPR nanoparticles, followed by standard chemotherapy (see Figure 2). Tumours treated with the combination showed arrested growth compared to those receiving chemotherapy alone. The results demonstrated that modest levels of gene editing – substantially lower than the near-complete editing often sought in other applications – were sufficient to restore chemosensitivity.
"We demonstrate that 20%-40% gene editing activity is sufficient to improve response to chemotherapy in animal models," the authors stated, highlighting the clinical relevance of achievable editing levels.
The findings were replicated in patient-derived tumour models naturally harbouring the R34G mutation, where comparable editing efficiency and mRNA localisation were achieved, supporting the approach's applicability across diverse genetic backgrounds.
The work establishes a framework for developing CRISPR-directed gene editing as an adjunct therapy to enhance standard cancer treatments. By strategically targeting a driver of drug resistance rather than attempting complete tumour elimination through gene editing alone, the approach could enable patients to tolerate lower chemotherapy doses whilst maintaining therapeutic efficacy – potentially reducing side effects and extending treatment duration.
The study was led by Kelly Banas and Eric Kmiec at the Gene Editing Institute at ChristianaCare Health System in Delaware. The research was published in Molecular Therapy Oncology on 13 November 2025.
To get more CRISPR Medicine News delivered to your inbox, sign up to the free weekly CMN Newsletter here.
Tags
ArticleCMN HighlightsNewsStickyIn vivoLipid-based nanoparticleCancerLung CancerCas9
CLINICAL TRIALS
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.
Sponsors:
Base Therapeutics (Shanghai) Co., Ltd.







